CRISPRone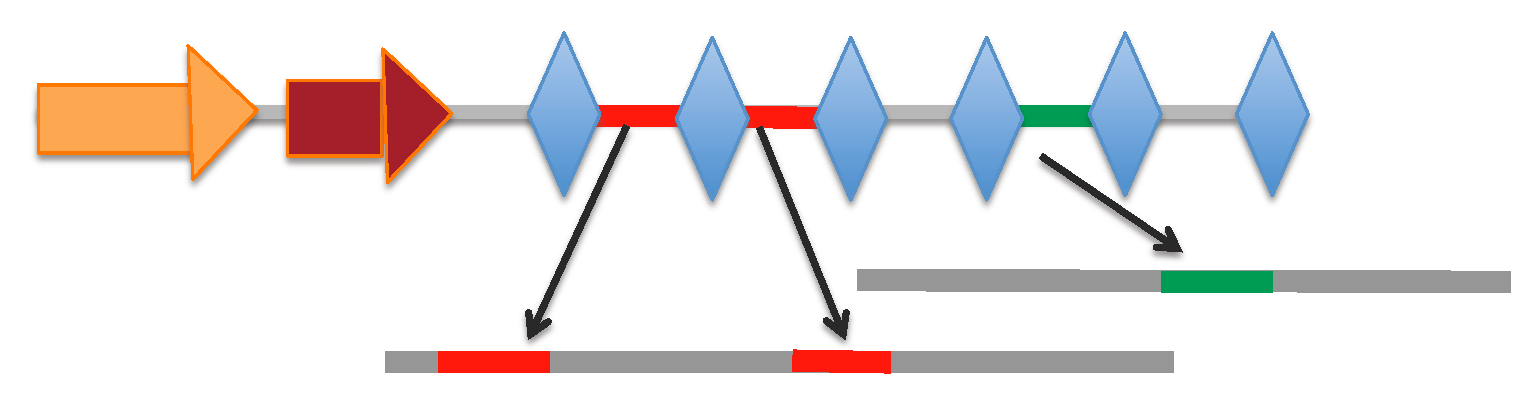
CRISPRone report for GCA_002154915.1_ASM215491v1
♦ Seq 1: MUKO01000003.1 Pseudomonas aeruginosa strain SCH_ABX19 ABX19_contig000003, whole genome shotgun sequence
⇒Summary: seq-len = 546205 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 336957-337586 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 2: MUKO01000008.1 Pseudomonas aeruginosa strain SCH_ABX19 ABX19_contig000008, whole genome shotgun sequence
⇒Summary: seq-len = 228548 bp; # of CRISPR array = 2; # of cas gene = 6 (type I) 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒CRISPR arrays :
[Locus range 181172-181620 bp]: array | spacers
[Locus range 190193-190339 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)



[Locus range 181172-181620 bp]: array | spacers
[Locus range 190193-190339 bp]: array | spacers
