CRISPRone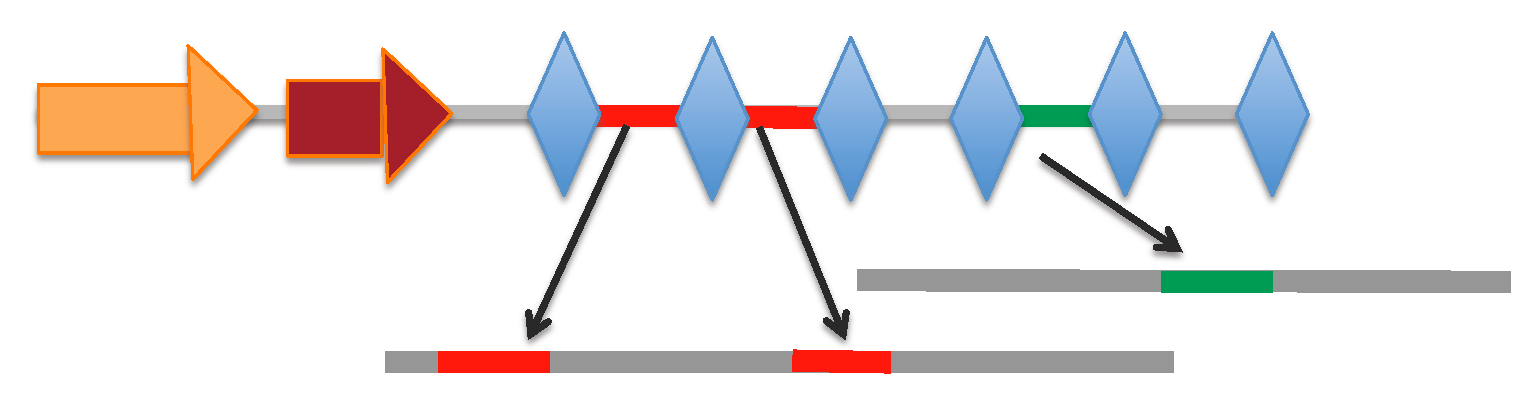
CRISPRone report for GCA_003633495.1_ASM363349v1
♦ Seq 1: QZXH01000025.1 Pseudomonas aeruginosa strain ENV-567 PA4_G3_NODE_23_length_93529_cov_37.7445_ID_45, whole genome shotgun sequence
⇒Summary: seq-len = 93529 bp; # of CRISPR array = 0; # of cas gene = 2 NA 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 2: QZXH01000063.1 Pseudomonas aeruginosa strain ENV-567 PA4_G3_NODE_39_length_47477_cov_35.3748_ID_77, whole genome shotgun sequence
⇒Summary: seq-len = 47477 bp; # of CRISPR array = 1; # of cas gene = 6 (type I) 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒CRISPR arrays :
[Locus range 38424-38992 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 3: QZXH01000033.1 Pseudomonas aeruginosa strain ENV-567 PA4_G3_NODE_63_length_1364_cov_39.6353_ID_125, whole genome shotgun sequence
⇒Summary: seq-len = 1364 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 139-1249 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 4: QZXH01000035.1 Pseudomonas aeruginosa strain ENV-567 PA4_G3_NODE_38_length_48743_cov_42.6618_ID_75, whole genome shotgun sequence
⇒Summary: seq-len = 48743 bp; # of CRISPR array = 0; # of cas gene = 3 (type I) 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 5: QZXH01000048.1 Pseudomonas aeruginosa strain ENV-567 PA4_G3_NODE_9_length_217199_cov_38.4165_ID_17, whole genome shotgun sequence
⇒Summary: seq-len = 217199 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 85182-86470 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
 )
)
[Locus range 139-1249 bp]: array | spacers
