CRISPRone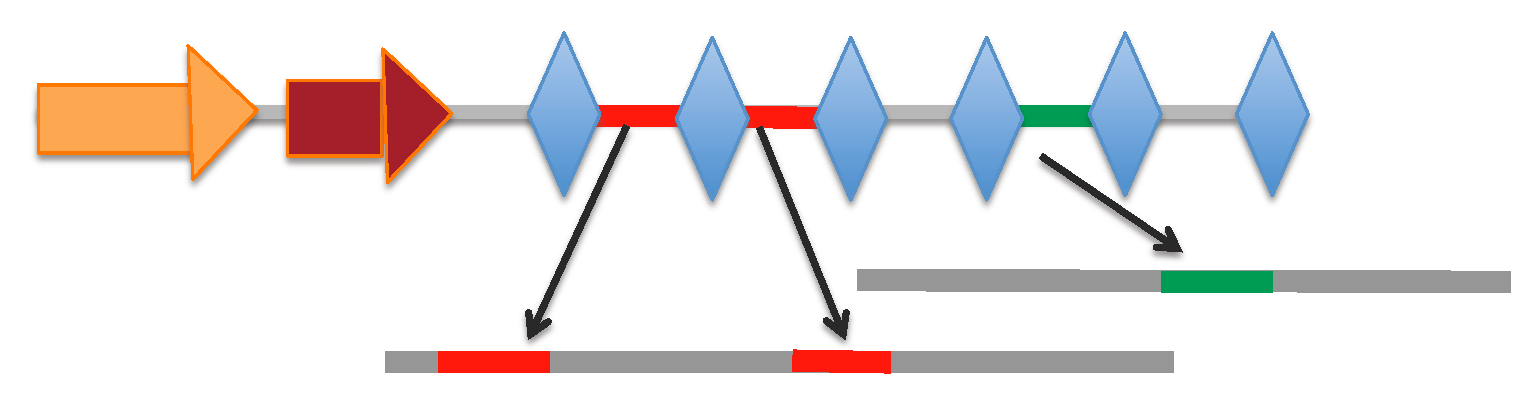
CRISPRone report for GCA_012987005.1_ASM1298700v1
♦ Seq 1: JAAQZH010000011.1 Pseudomonas aeruginosa strain WS 5022 11_247346_49.0127, whole genome shotgun sequence
⇒Summary: seq-len = 247346 bp; # of CRISPR array = 2; # of cas gene = 6 (type I) 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒CRISPR arrays :
[Locus range 207726-208294 bp]: array | spacers
[Locus range 217236-217924 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 2: JAAQZH010000014.1 Pseudomonas aeruginosa strain WS 5022 14_197935_58.7894, whole genome shotgun sequence
⇒Summary: seq-len = 197935 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 29235-29562 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
 )
)
[Locus range 29235-29562 bp]: array | spacers
