CRISPRone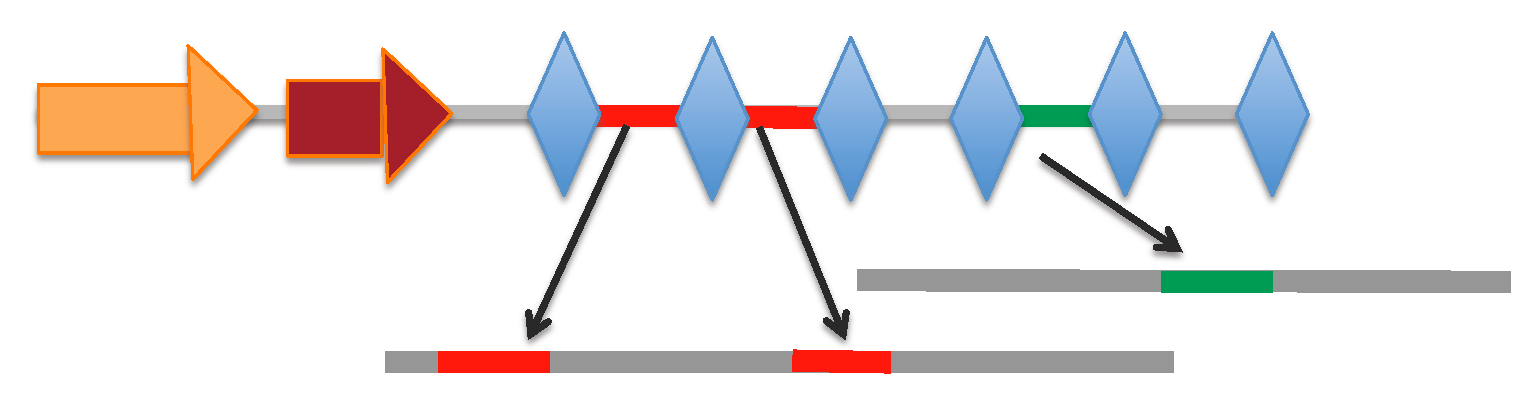
CRISPRone report for GCA_900007545.1_ASM90000754v1
♦ Seq 1: CZWK01000019.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_19_length_79896_cov_15.7051_ID_39, whole genome shotgun sequence
⇒Summary: seq-len = 79896 bp; # of CRISPR array = 2; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 23451-23740 bp]: array | spacers
[Locus range 25605-25961 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 2: CZWK01000046.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_46_length_31812_cov_12.484_ID_93, whole genome shotgun sequence
⇒Summary: seq-len = 31812 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 30083-31040 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 3: CZWK01000004.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_4_length_141897_cov_16.779_ID_7, whole genome shotgun sequence
⇒Summary: seq-len = 141897 bp; # of CRISPR array = 1; # of cas gene = 10 (type I) 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒CRISPR arrays :
[Locus range 26346-27964 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 4: CZWK01000032.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_32_length_48085_cov_13.3537_ID_65, whole genome shotgun sequence
⇒Summary: seq-len = 48085 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 23471-23765 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 5: CZWK01000021.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_21_length_73081_cov_14.8143_ID_43, whole genome shotgun sequence
⇒Summary: seq-len = 73081 bp; # of CRISPR array = 0; # of cas gene = 6 (type I, III) 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 6: CZWK01000001.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_1_length_184781_cov_14.6095_ID_1, whole genome shotgun sequence
⇒Summary: seq-len = 184781 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 5864-6550 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 7: CZWK01000052.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_52_length_27163_cov_9.64014_ID_105, whole genome shotgun sequence
⇒Summary: seq-len = 27163 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 2043-2137 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 8: CZWK01000009.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_9_length_96396_cov_15.6368_ID_17, whole genome shotgun sequence
⇒Summary: seq-len = 96396 bp; # of CRISPR array = 1; # of cas gene = 5 (type I) 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒CRISPR arrays :
[Locus range 69345-69967 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 9: CZWK01000049.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_49_length_28644_cov_11.3066_ID_99, whole genome shotgun sequence
⇒Summary: seq-len = 28644 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 26077-26703 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 10: CZWK01000056.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_56_length_21143_cov_12.1556_ID_113, whole genome shotgun sequence
⇒Summary: seq-len = 21143 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 19532-20356 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
 )
)
[Locus range 30083-31040 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 3: CZWK01000004.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_4_length_141897_cov_16.779_ID_7, whole genome shotgun sequence
⇒Summary: seq-len = 141897 bp; # of CRISPR array = 1; # of cas gene = 10 (type I) 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒CRISPR arrays :
[Locus range 26346-27964 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 4: CZWK01000032.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_32_length_48085_cov_13.3537_ID_65, whole genome shotgun sequence
⇒Summary: seq-len = 48085 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 23471-23765 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 5: CZWK01000021.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_21_length_73081_cov_14.8143_ID_43, whole genome shotgun sequence
⇒Summary: seq-len = 73081 bp; # of CRISPR array = 0; # of cas gene = 6 (type I, III) 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 6: CZWK01000001.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_1_length_184781_cov_14.6095_ID_1, whole genome shotgun sequence
⇒Summary: seq-len = 184781 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 5864-6550 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 7: CZWK01000052.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_52_length_27163_cov_9.64014_ID_105, whole genome shotgun sequence
⇒Summary: seq-len = 27163 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 2043-2137 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 8: CZWK01000009.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_9_length_96396_cov_15.6368_ID_17, whole genome shotgun sequence
⇒Summary: seq-len = 96396 bp; # of CRISPR array = 1; # of cas gene = 5 (type I) 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒CRISPR arrays :
[Locus range 69345-69967 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 9: CZWK01000049.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_49_length_28644_cov_11.3066_ID_99, whole genome shotgun sequence
⇒Summary: seq-len = 28644 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 26077-26703 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 10: CZWK01000056.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_56_length_21143_cov_12.1556_ID_113, whole genome shotgun sequence
⇒Summary: seq-len = 21143 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 19532-20356 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
 )
)
[Locus range 23471-23765 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 5: CZWK01000021.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_21_length_73081_cov_14.8143_ID_43, whole genome shotgun sequence
⇒Summary: seq-len = 73081 bp; # of CRISPR array = 0; # of cas gene = 6 (type I, III) 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 6: CZWK01000001.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_1_length_184781_cov_14.6095_ID_1, whole genome shotgun sequence
⇒Summary: seq-len = 184781 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 5864-6550 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 7: CZWK01000052.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_52_length_27163_cov_9.64014_ID_105, whole genome shotgun sequence
⇒Summary: seq-len = 27163 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 2043-2137 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 8: CZWK01000009.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_9_length_96396_cov_15.6368_ID_17, whole genome shotgun sequence
⇒Summary: seq-len = 96396 bp; # of CRISPR array = 1; # of cas gene = 5 (type I) 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒CRISPR arrays :
[Locus range 69345-69967 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 9: CZWK01000049.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_49_length_28644_cov_11.3066_ID_99, whole genome shotgun sequence
⇒Summary: seq-len = 28644 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 26077-26703 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 10: CZWK01000056.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_56_length_21143_cov_12.1556_ID_113, whole genome shotgun sequence
⇒Summary: seq-len = 21143 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 19532-20356 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
 )
)
[Locus range 2043-2137 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 8: CZWK01000009.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_9_length_96396_cov_15.6368_ID_17, whole genome shotgun sequence
⇒Summary: seq-len = 96396 bp; # of CRISPR array = 1; # of cas gene = 5 (type I) 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒CRISPR arrays :
[Locus range 69345-69967 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 9: CZWK01000049.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_49_length_28644_cov_11.3066_ID_99, whole genome shotgun sequence
⇒Summary: seq-len = 28644 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 26077-26703 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 10: CZWK01000056.1 Peptoclostridium difficile isolate VL_0002 genome assembly, contig: NODE_56_length_21143_cov_12.1556_ID_113, whole genome shotgun sequence
⇒Summary: seq-len = 21143 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 19532-20356 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
 )
)
[Locus range 26077-26703 bp]: array | spacers
