CRISPRone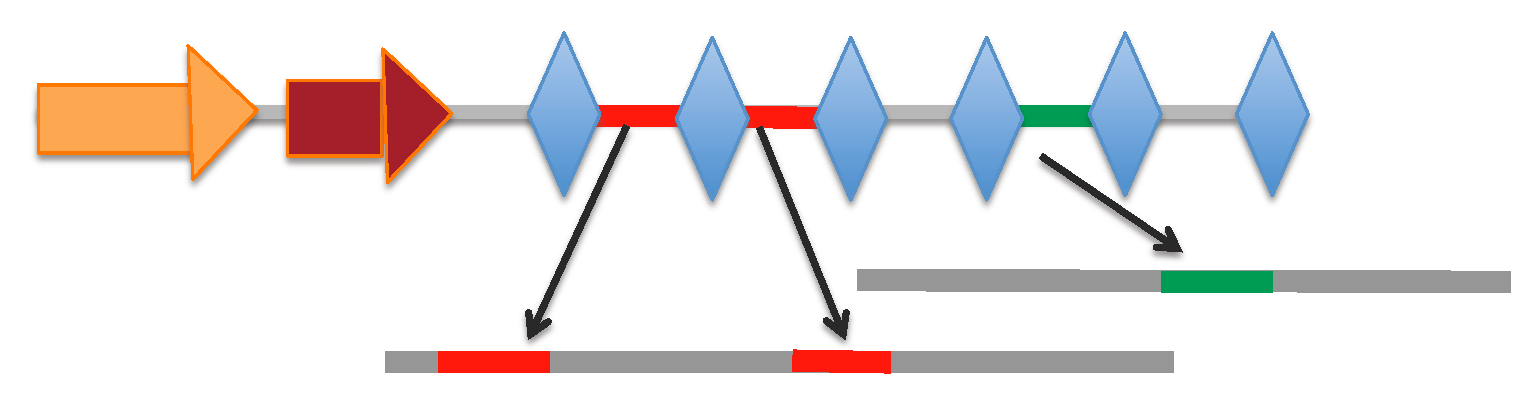
CRISPRone report for GCA_900147245.1_398
♦ Seq 1: FRSQ01000213.1 Pseudomonas aeruginosa isolate 398 genome assembly, contig: 398.NODE_295_length_17201_cov_52.611012, whole genome shotgun sequence
⇒Summary: seq-len = 17243 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 260-1009 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 2: FRSQ01000018.1 Pseudomonas aeruginosa isolate 398 genome assembly, contig: 398.NODE_19_length_36380_cov_52.522568, whole genome shotgun sequence
⇒Summary: seq-len = 36422 bp; # of CRISPR array = 2; # of cas gene = 6 (type I) 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒CRISPR arrays :
[Locus range 258-826 bp]: array | spacers
[Locus range 9750-10857 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)



[Locus range 258-826 bp]: array | spacers
[Locus range 9750-10857 bp]: array | spacers
