CRISPRone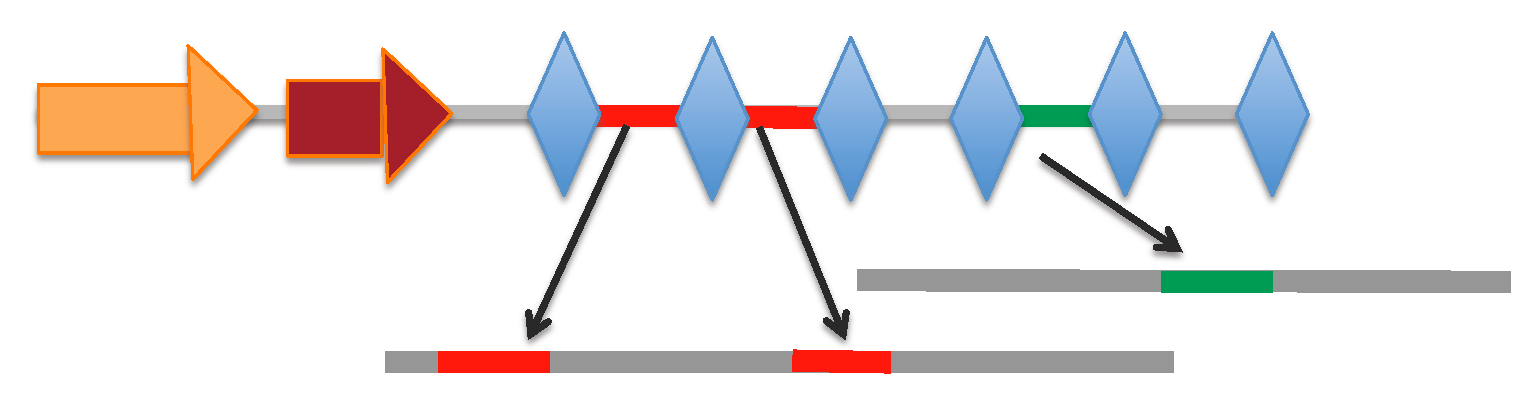
CRISPRone report for GCA_901005945.1_8140_6_2
♦ Seq 1: CAAKAM010000002.1 Clostridioides difficile strain LSHTM-2 genome assembly, contig: ERS139975SCcontig000002, whole genome shotgun sequence
⇒Summary: seq-len = 376728 bp; # of CRISPR array = 1; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 194180-194275 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 2: CAAKAM010000001.1 Clostridioides difficile strain LSHTM-2 genome assembly, contig: ERS139975SCcontig000001, whole genome shotgun sequence
⇒Summary: seq-len = 528650 bp; # of CRISPR array = 5; # of cas gene = 0 (Questionable CRISPR )
)
⇒Cas loci & CRISPR arrays: gff file 
⇒CRISPR arrays :
[Locus range 28790-28951 bp]: array | spacers
[Locus range 138210-138830 bp]: array | spacers
[Locus range 229679-229906 bp]: array | spacers
[Locus range 232573-232734 bp]: array | spacers
[Locus range 364123-364543 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
♦ Seq 3: CAAKAM010000003.1 Clostridioides difficile strain LSHTM-2 genome assembly, contig: ERS139975SCcontig000003, whole genome shotgun sequence
⇒Summary: seq-len = 345164 bp; # of CRISPR array = 1; # of cas gene = 10 (type I, III) 
⇒Cas loci & CRISPR arrays: gff file 
⇒Predicted cas genes: protein | gene 
⇒CRISPR arrays :
[Locus range 247009-247234 bp]: array | spacers
⇒Miscellaneous: suspicious cas genes and/or false-CRISPRs (link)
 )
)
[Locus range 28790-28951 bp]: array | spacers
[Locus range 138210-138830 bp]: array | spacers
[Locus range 229679-229906 bp]: array | spacers
[Locus range 232573-232734 bp]: array | spacers
[Locus range 364123-364543 bp]: array | spacers
