myRT
Demonstration examples
- Plasmids carrying a group II intron reverse transcriptase adjacent to IMP resistance genes [see myRT prediction]
Enterobacter hormaechei, a gram-negative bacterium, is a drug resistant opportunistic infectious pathogen. A clinical isolate of E. hormaechei L51 produces , and class 1 integron carrying gene cassette in Multi-Drug resistant (MDR) region of pEHZJ1, mega conjugative IncHI2/2A plasmid of E. hormaechei L51, contains blaIMP-26, and a group II intron reverse transcriptase . pIMP-PH114 plasmid in Klebsiella pneumoniae Strain harbouring blaIMP-4 is found to be closely related to other Multi drug-Resistant IncA/C2 Plasmids. The spread of these plasmids that carry IMP resistance genes is alarming, and is worth studying as infections caused by multi drug resistant bacteria kills many people each year (ref1, ref2 ).
- A Transposon encoding a Group II intron Reverse Transcriptase [see myRT prediction ]
Transposons are mobile genetic elements (MGEs) that can change their locations in the host genome either via a cut-and-paste mechanism in type 1, or an RNA intermediate reverse transcribed into cDNA in type 2 (retrotransposons). Most transposases belong to the DDE superfamily, however some transposons encode transposases that are homologous to HUH, tyrosine recombinase, or serine integrases/invertases. Some bacterial viruses encode transposases to integrate their viral genome into the host. Some transposases (e.g. ISLdr1 family transposase in Legionella drancourtii LLAP12) encode a reverse transcriptase ( ref1 , ref2 , ref3 )
- Transposases in the neighborhood of DGR reverse transcriptases
A recent study (ref) suggested that Target genes in Diversity generating retroelemets (DGRs), may have been proliferated from a common ancestral gene by means of transposition and duplication. They searched the genomic neighborhood ( > 5000 bps) of different target genes of DGR systems to find transposases in 21 genomes with DGR systems, among which Trichodesmium erythraeum IMS 101 has the highest number of transposases in the genomic neighborhood of target genes. And, IS200/IS605 with peel and paste transposition mechanism is the most common transposase within the neighborhood of these Target genes, suggesting that these transposase genes may play a role in target gene dispersal (ref).
Genome putative RT myRT results Transposase(s) Acaryochloris marina MBIC11017 ABW25404.1 myRT-prediction ABW25403.1 (DDE_Tnp_1_5, COG3293 domains) Anabaena sp. 90 plasmid pANA02 AFW97271.1 myRT-prediction AFW97274.1 (YhhI domain), AFW97275.1 (DDE_5 domain) Scytonema sp. HK-05 EAZ90090.1 myRT-prediction OKH60567.1 (IS200/IS605 family; RAYT domain --REP element-mobilizing transposase RayT-- ) AP018194.1 Scytonema sp. HK-05 BAY45961.1 myRT-prediction BAY45959.1 (IS200-family; RAYT domain)
- Casposons containing a Retrons reverse transcriptase
Casposons are bacterial and archaeal mobile genetic elements, that are able to self replicate. The recombinase (casposase) in these transposons, provides them with integration into the host genomes, and is homologous to Cas1 endonuclease, which is involved in CISPR-Cas immunity. Phylogenetic analysis suggests that casposons might have played a crucial role in emergence of adaptive immunity in CRISPR-Cas systems. Only a few casposons do have a reverse transcriptase; for instance, the casposons in Methanosarcina sp. (2.H.T.1A.3, 2.H.T.1A.6, 2.H.T.1A.8, and 2.H.T.1A.15) contain a putative reverse transcriptase which is adjacent to Cas1 endonuclease (ref1, ref2, ref3).
Genome myRT results Methanosarcina sp. 2.H.T.1A.3 myRT-prediction Methanosarcina sp. 2.H.T.1A.6 myRT-prediction Methanosarcina sp. 2.H.T.1A.8 myRT-prediction Methanosarcina sp. 2.H.T.1A.15 myRT-prediction
- Group II introns RTs in Bacillus cereus group (ref)
[see myRT prediction results for Group II introns RTs in Bacillus cereus ATCC 10987 (B.c.I1(b),B.c.I2(c), B.c.I3) and plasmid pBc10987 (B.c.I4, B.c.I1(c), B.c.I5, B.c.I2(b))]
[see myRT prediction for Group II introns RTs in Bacillus anthracis str. A2012 plasmid pXO1 (B.a.I1, B.a.I2)]
[see myRT visualization for Group II introns RTs in Bacillus cereus ATCC 14579 (B.c.I1(a))]
[see myRT visualization for Bacillus cereus Q1 with two group II introns RTs]
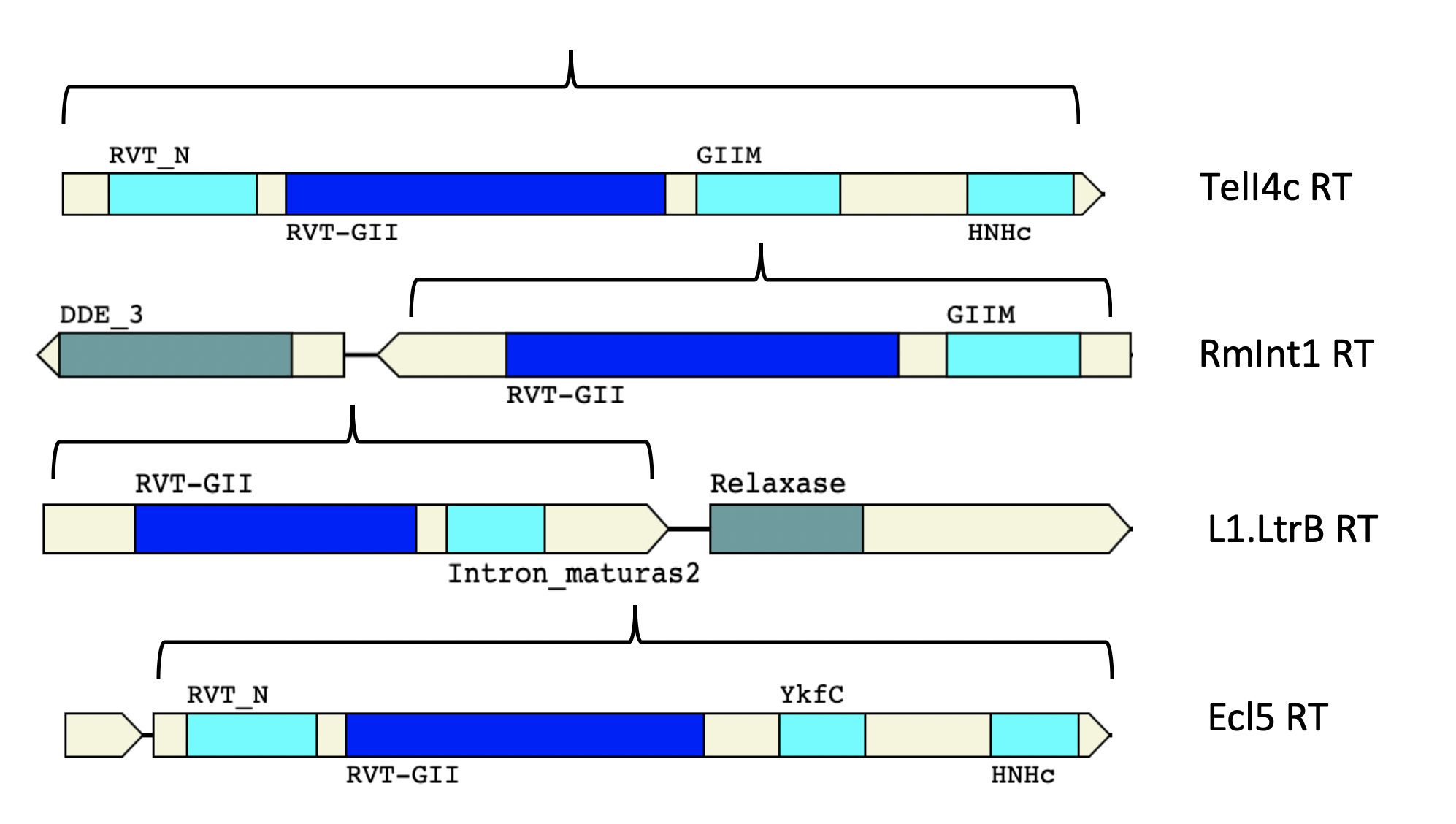
- Different Group II introns RTs used as Thermotargetron and Targetrons
- Reverse Transcriptases in Skin Metagenome (ref) [see myRT prediction]