myDGR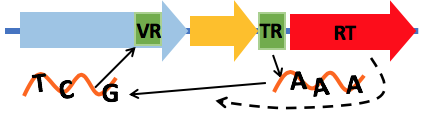
Putative DGR systems surveyed in:
Putative DGR systems in Core Set
| Index | Accession Number | Organism | Phylum/Class | DGR set | Type | DGR |
|---|---|---|---|---|---|---|
| 1 | AGVU01000027.1 | Fluoribacter dumoffii NY 23 | Proteobacteria/Gammaproteobacteria | core set | plasmid | myDGR prediction |
| 2 | NC_009494.2 | Legionella pneumophila str. Corby | Proteobacteria/Gammaproteobacteria | core set | chromosome | myDGR prediction |
| 3 | NC_018011.1 | Alistipes finegoldii DSM 17242 | Bacteroidetes/Bacteroidia | core set | chromosome | myDGR prediction |
| 4 | CAPH01000002.1 | Alistipes sp. AP11 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 5 | NZ_AJIN01000036.1 | Prevotella sp. oral taxon 306 str. F0472 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 6 | NC_004663.1 | Bacteroides thetaiotaomicron VPI-5482 | Bacteroidetes/Bacteroidia | core set | chromosome | myDGR prediction |
| 7 | CAHI01000003.1 | Alistipes sp. JC50 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 8 | NZ_AHHG01000042.1 | Elizabethkingia anophelis Ag1 | Bacteroidetes/Flavobacteriia | core set | WGS | myDGR prediction |
| 9 | CP003283.1 | Ornithobacterium rhinotracheale DSM 15997 | Bacteroidetes/Flavobacteriia | core set | chromosome | myDGR prediction |
| 10 | AAYH02000035.1 | Bacteroides uniformis ATCC 8492 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 11 | AGXT01000008.1 | Bacteroides ovatus CL02T12C04 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 12 | NC_014033.1 | Prevotella ruminicola 23 | Bacteroidetes/Bacteroidia | core set | chromosome | myDGR prediction |
| 13 | ADJS01000105.1 | uncultured Alistipes sp. | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 14 | AGXW01000012.1 | Bacteroides finegoldii CL09T03C10 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 15 | NZ_ADMO01000036.1 | Bacteroides ovatus SD CMC 3f | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 16 | FP929033.1 | Bacteroides xylanisolvens XB1A | Bacteroidetes/Bacteroidia | core set | chromosome | myDGR prediction |
| 17 | ACPP01000012.1 | Bacteroides sp. 2_1_16 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 18 | FP929063.1 | Candidatus Magnetobacterium bavaricum | Nitrospirae/Nitrospira | core set | chromosome | myDGR prediction |
| 19 | AMFJ01002400.1 | uncultured bacterium | Unidentified | core set | metagenome | myDGR prediction |
| 20 | NZ_CH902599.1 | Photobacterium angustum S14 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 21 | NZ_ADBD01000009.1 | Vibrio sp. RC586 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 22 | NC_009036.1 | Shewanella baltica OS155 | Proteobacteria/Gammaproteobacteria | core set | plasmid | myDGR prediction |
| 23 | NC_010814.1 | Geobacter lovleyi SZ | Proteobacteria/Deltaproteobacteria | core set | chromosome | myDGR prediction |
| 24 | ACDV01000044.1 | Stenotrophomonas sp. SKA14 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 25 | NZ_AJXW01000043.1 | Rhodanobacter thiooxydans LCS2 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 26 | NZ_AGFD01000005.1 | Thiorhodospira sibirica ATCC 700588 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 27 | FR695874.1 | uncultured Desulfobacterium sp. | Proteobacteria/Deltaproteobacteria | core set | metagenome | myDGR prediction |
| 28 | AMWB01060384.1 | bioreactor metagenome | Unidentified | core set | metagenome | myDGR prediction |
| 29 | AMWB01080710.1 | bioreactor metagenome | Unidentified | core set | metagenome | myDGR prediction |
| 30 | FP885895.1 | Ralstonia solanacearum CMR15 | Proteobacteria/Betaproteobacteria | core set | chromosome | myDGR prediction |
| 31 | CAFQ01000015.1 | Burkholderia cenocepacia H111 | Proteobacteria/Betaproteobacteria | core set | WGS | myDGR prediction |
| 32 | NZ_ABBM01001082.1 | Burkholderia thailandensis MSMB43 | Proteobacteria/Betaproteobacteria | core set | WGS | myDGR prediction |
| 33 | NC_012724.2 | Burkholderia glumae BGR1 | Proteobacteria/Betaproteobacteria | core set | chromosome | myDGR prediction |
| 34 | AMXB01000038.1 | Thauera sp. 27 | Proteobacteria/Betaproteobacteria | core set | WGS | myDGR prediction |
| 35 | NZ_AJWX01000011.1 | Pseudomonas sp. M47T1 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 36 | NZ_AKJH01000019.1 | Pseudomonas sp. GM67 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 37 | AJXF01000114.1 | Pseudomonas fluorescens NZI7 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 38 | NC_017911.1 | Pseudomonas fluorescens A506 | Proteobacteria/Gammaproteobacteria | core set | chromosome | myDGR prediction |
| 39 | NZ_CM001025.1 | Pseudomonas fluorescens WH6 | Proteobacteria/Gammaproteobacteria | core set | chromosome | myDGR prediction |
| 40 | NZ_AKJT01000025.1 | Pseudomonas sp. GM18 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 41 | NZ_AKIV01000010.1 | Polaromonas sp. CF318 | Proteobacteria/Betaproteobacteria | core set | WGS | myDGR prediction |
| 42 | NZ_AHHB01000028.1 | Janthinobacterium sp. PAMC 25724 | Proteobacteria/Betaproteobacteria | core set | WGS | myDGR prediction |
| 43 | CP002745.1 | Collimonas fungivorans Ter331 | Proteobacteria/Betaproteobacteria | core set | chromosome | myDGR prediction |
| 44 | CP001965.1 | Sideroxydans lithotrophicus ES-1 | Proteobacteria/Betaproteobacteria | core set | chromosome | myDGR prediction |
| 45 | CP002449.1 | Alicycliphilus denitrificans BC | Proteobacteria/Betaproteobacteria | core set | chromosome | myDGR prediction |
| 46 | NC_016002.1 | Pseudogulbenkiania sp. NH8B | Proteobacteria/Gammaproteobacteria | core set | chromosome | myDGR prediction |
| 47 | AFXK01000001.1 | Pseudomonas aeruginosa 213BR | Proteobacteria/Gammaproteobacteria | core set | chromosome | myDGR prediction |
| 48 | NZ_ANIR01000003.1 | Pseudomonas sp. M1 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 49 | NZ_AMWJ01000046.1 | Pseudomonas putida CSV86 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 50 | AP007255.1 | Magnetospirillum magneticum AMB-1 | Proteobacteria/Alphaproteobacteria | core set | chromosome | myDGR prediction |
| 51 | NC_007626.1 | Magnetospirillum magneticum AMB-1 | Proteobacteria/Alphaproteobacteria | core set | chromosome | myDGR prediction |
| 52 | NC_008639.1 | Chlorobium phaeobacteroides DSM 266 | Chlorobi/Chlorobia | core set | chromosome | myDGR prediction |
| 53 | NC_011060.1 | Pelodictyon phaeoclathratiforme BU-1 | Chlorobi/Chlorobia | core set | chromosome | myDGR prediction |
| 54 | CP001965.1 | Sideroxydans lithotrophicus ES-1 | Proteobacteria/Betaproteobacteria | core set | chromosome | myDGR prediction |
| 55 | CP000828.1 | Acaryochloris marina MBIC11017 | Cyanobacteria/Oscillatoriophycideae | core set | chromosome | myDGR prediction |
| 56 | ALWB01000040.1 | Pseudanabaena biceps PCC 7429 | Cyanobacteria/Oscillatoriophycideae | core set | WGS | myDGR prediction |
| 57 | NZ_CAFN01000268.1 | Arthrospira sp. PCC 8005 | Cyanobacteria/Cyanophyceae | core set | WGS | myDGR prediction |
| 58 | NZ_AAXW01000046.1 | Cyanothece sp. CCY0110 | Cyanobacteria/Cyanophyceae | core set | WGS | myDGR prediction |
| 59 | NC_010628.1 | Nostoc punctiforme PCC 73102 | Cyanobacteria/Cyanophyceae | core set | chromosome | myDGR prediction |
| 60 | BA000019.2 | Nostoc sp. PCC 7120 | Cyanobacteria/Cyanophyceae | core set | chromosome | myDGR prediction |
| 61 | NZ_AAVW01000008.1 | Nodularia spumigena CCY9414 | Cyanobacteria/Cyanophyceae | core set | WGS | myDGR prediction |
| 62 | ALVJ01000051.1 | Calothrix sp. PCC 7103 | Cyanobacteria/Cyanophyceae | core set | WGS | myDGR prediction |
| 63 | ALVJ01000038.1 | Calothrix sp. PCC 7103 | Cyanobacteria/Cyanophyceae | core set | WGS | myDGR prediction |
| 64 | ALVJ01000040.1 | Calothrix sp. PCC 7103 | Cyanobacteria/Cyanophyceae | core set | WGS | myDGR prediction |
| 65 | CP000393.1 | Trichodesmium erythraeum IMS101 | Cyanobacteria/Oscillatoriophycideae | core set | chromosome | myDGR prediction |
| 66 | ALVW01000006.1 | Mastigocladopsis repens PCC 10914 | Cyanobacteria/Stigonematales | core set | WGS | myDGR prediction |
| 67 | AJLK01000113.1 | Fischerella muscicola PCC 7414 | Cyanobacteria/Cyanophyceae | core set | WGS | myDGR prediction |
| 68 | CP003287.1 | Anabaena sp. 90 | Cyanobacteria/Nostocales | core set | plasmid | myDGR prediction |
| 69 | AEPQ01000184.1 | Moorea producens 3L | Cyanobacteria/Cyanophyceae | core set | WGS | myDGR prediction |
| 70 | ALVK01000015.1 | Pseudanabaena sp. PCC 6802 | Cyanobacteria/Oscillatoriophycideae | core set | WGS | myDGR prediction |
| 71 | CP003946.1 | Leptolyngbya sp. PCC 7376 | Cyanobacteria/Cyanophyceae | core set | chromosome | myDGR prediction |
| 72 | ALVN01000008.1 | Leptolyngbya sp. PCC 7375 | Cyanobacteria/Cyanophyceae | core set | WGS | myDGR prediction |
| 73 | ALVN01000008.1 | Leptolyngbya sp. PCC 7375 | Cyanobacteria/Cyanophyceae | core set | WGS | myDGR prediction |
| 74 | ANKN01000155.1 | Prochlorothrix hollandica PCC 9006 | Cyanobacteria/Prochlorales | core set | WGS | myDGR prediction |
| 75 | ALVR01000007.1 | Spirulina subsalsa PCC 9445 | Cyanobacteria/Chroobacteria | core set | WGS | myDGR prediction |
| 76 | NZ_AAXW01000031.1 | Cyanothece sp. CCY0110 | Cyanobacteria/Cyanophyceae | core set | WGS | myDGR prediction |
| 77 | NC_008312.1 | Trichodesmium erythraeum IMS101 | Cyanobacteria/Oscillatoriophycideae | core set | chromosome | myDGR prediction |
| 78 | AJWF01000011.1 | Anabaena sp. PCC 7108 | Cyanobacteria/Nostocales | core set | WGS | myDGR prediction |
| 79 | NC_003272.1 | Nostoc sp. PCC 7120 | Cyanobacteria/Cyanophyceae | core set | chromosome | myDGR prediction |
| 80 | ANFJ01000002.1 | Microchaete sp. PCC 7126 | Cyanobacteria/Cyanophyceae | core set | WGS | myDGR prediction |
| 81 | AESD01001019.1 | Crocosphaera watsonii WH 0003 | Cyanobacteria/Cyanophyceae | core set | WGS | myDGR prediction |
| 82 | ANKO01000117.1 | Oscillatoria sp. PCC 10802 | Cyanobacteria/Cyanophyceae | core set | WGS | myDGR prediction |
| 83 | NZ_AAVW01000038.1 | Nodularia spumigena CCY9414 | Cyanobacteria/Cyanophyceae | core set | WGS | myDGR prediction |
| 84 | NC_013194.1 | Candidatus Accumulibacter phosphatis clade IIA str. UW-1 | Proteobacteria/Betaproteobacteria | core set | chromosome | myDGR prediction |
| 85 | NZ_AFWS02000055.1 | Thiorhodovibrio sp. 970 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 86 | NC_014664.1 | Rhodomicrobium vannielii ATCC 17100 | Proteobacteria/Alphaproteobacteria | core set | chromosome | myDGR prediction |
| 87 | NC_015510.1 | Haliscomenobacter hydrossis DSM 1100 | Bacteroidetes/Bacteroidia | core set | chromosome | myDGR prediction |
| 88 | NS_000195.1 | Candidatus Cloacamonas acidaminovorans | Unclassified bacterium | core set | metagenome | myDGR prediction |
| 89 | NC_011061.1 | Prosthecochloris aestuarii DSM 271 | Chlorobi/Chlorobia | core set | plasmid | myDGR prediction |
| 90 | NZ_AFWU01000006.1 | Marichromatium purpuratum 984 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 91 | NC_002967.9 | Treponema denticola ATCC 35405 | Spirochaetes/Spirochaetia | core set | chromosome | myDGR prediction |
| 92 | NC_011060.1 | Pelodictyon phaeoclathratiforme BU-1 | Chlorobi/Chlorobia | core set | chromosome | myDGR prediction |
| 93 | NC_007514.1 | Chlorobium chlorochromatii CaD3 | Chlorobi/Chlorobia | core set | chromosome | myDGR prediction |
| 94 | AMQJ01000472.1 | sediment metagenome | Unidentified | core set | metagenome | myDGR prediction |
| 95 | NC_013194.1 | Candidatus Accumulibacter phosphatis clade IIA str. UW-1 | Proteobacteria/Betaproteobacteria | core set | chromosome | myDGR prediction |
| 96 | AATN01000314.1 | metagenome | Unidentified | core set | metagenome | myDGR prediction |
| 97 | NZ_CM001488.1 | Desulfobacter postgatei 2ac9 | Proteobacteria/Deltaproteobacteria | core set | chromosome | myDGR prediction |
| 98 | NZ_AFWT01000016.1 | Thiorhodococcus drewsii AZ1 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 99 | NZ_ABVK02000003.1 | Thermus aquaticus Y51MC23 | Deinococcus-Thermus/Deinococci | core set | WGS | myDGR prediction |
| 100 | NZ_ABEA02000065.1 | Diplosphaera colitermitum TAV2 | Verrucomicrobia/Opitutae | core set | WGS | myDGR prediction |
| 101 | CBDL010000055.1 | Clostridium sp. CAG:575 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 102 | CAXX010000057.1 | Acholeplasma sp. CAG:878 | Tenericutes/Mollicutes | core set | WGS | myDGR prediction |
| 103 | CBDJ010000043.1 | Firmicutes bacterium CAG:582 | Firmicutes/- | core set | WGS | myDGR prediction |
| 104 | CAZG010000040.1 | Clostridium sp. CAG:793 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 105 | CBGO010000007.1 | Clostridium sp. CAG:798 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 106 | AMWB01059641.1 | bioreactor metagenome | Unidentified | core set | metagenome | myDGR prediction |
| 107 | JQ680369.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 108 | CAPH01000015.1 | Alistipes sp. AP11 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 109 | NZ_DS499577.1 | Alistipes putredinis DSM 17216 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 110 | NZ_DS981430.1 | Bacteroides coprocola DSM 17136 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 111 | ADJS01003203.1 | uncultured Bacteroides sp. | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 112 | NZ_AMSG01000002.1 | Galbibacter sp. ck-I2-15 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 113 | AKZJ01000008.1 | Pedobacter arcticus A12 | Bacteroidetes/Sphingobacteria | core set | WGS | myDGR prediction |
| 114 | FP929032.1 | Alistipes shahii WAL 8301 | Bacteroidetes/Bacteroidia | core set | chromosome | myDGR prediction |
| 115 | ABZX01000035.1 | Bacteroides fragilis 3_1_12 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 116 | AMWB01221839.1 | bioreactor metagenome | Unidentified | core set | metagenome | myDGR prediction |
| 117 | NC_009615.1 | Parabacteroides distasonis ATCC 8503 | Bacteroidetes/Bacteroidia | core set | chromosome | myDGR prediction |
| 118 | NZ_EQ973486.1 | Bacteroides cellulosilyticus DSM 14838 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 119 | AGXT01000002.1 | Bacteroides ovatus CL02T12C04 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 120 | JQ680372.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 121 | NC_015160.1 | Odoribacter splanchnicus DSM 20712 | Bacteroidetes/Bacteroidia | core set | chromosome | myDGR prediction |
| 122 | NZ_ABZX01000038.1 | Bacteroides fragilis 3_1_12 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 123 | NC_012782.1 | Eubacterium eligens ATCC 27750 | Firmicutes/Clostridia | core set | plasmid | myDGR prediction |
| 124 | ACWP01000027.1 | Lachnospiraceae bacterium 3_1_46FAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 125 | JQ680362.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 126 | NZ_GG774705.1 | Bacteroides sp. 1_1_14 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 127 | AGXW01000002.1 | Bacteroides finegoldii CL09T03C10 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 128 | NZ_GL945090.1 | Bacteroides sp. 1_1_30 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 129 | NZ_ADKO01000109.1 | Bacteroides vulgatus PC510 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 130 | AGXM01000015.1 | Bacteroides fragilis CL07T00C01 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 131 | ABQC02000022.1 | Bacteroides plebeius DSM 17135 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 132 | NZ_GL635657.1 | Bacteroides sp. 3_1_40A | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 133 | NZ_EQ973215.1 | Bacteroides fragilis 3_1_12 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 134 | AEQO01000177.1 | Prevotella salivae DSM 15606 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 135 | AGXJ01000009.1 | Bacteroides dorei CL02T12C06 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 136 | AGXU01000044.1 | Bacteroides ovatus CL03T12C18 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 137 | AGZQ01000025.1 | Parabacteroides merdae CL03T12C32 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 138 | ACKS01000095.1 | Prevotella bergensis DSM 17361 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 139 | NZ_JH114142.1 | Prevotella sp. C561 | Bacteroidetes/Bacteroidia | core set | WGS | myDGR prediction |
| 140 | ADJS01003586.1 | uncultured Bifidobacterium sp. | Actinobacteria/Actinobacteria | core set | WGS | myDGR prediction |
| 141 | AP010890.1 | Bifidobacterium longum subsp. infantis 157F | Actinobacteria/Actinobacteria | core set | chromosome | myDGR prediction |
| 142 | ADJT01002331.1 | uncultured Bifidobacterium sp. | Actinobacteria/Actinobacteria | core set | WGS | myDGR prediction |
| 143 | NZ_AABM02000005.1 | Bifidobacterium longum DJO10A | Actinobacteria/Actinobacteria | core set | WGS | myDGR prediction |
| 144 | FP929034.1 | Bifidobacterium longum subsp. longum F8 | Actinobacteria/Actinobacteria | core set | chromosome | myDGR prediction |
| 145 | NZ_AAVN02000001.1 | Collinsella aerofaciens ATCC 25986 | Actinobacteria/Actinobacteria | core set | WGS | myDGR prediction |
| 146 | FP929038.1 | Coprococcus catus GD/7 | Firmicutes/Clostridia | core set | chromosome | myDGR prediction |
| 147 | FP929046.1 | Faecalibacterium prausnitzii SL3/3 | Firmicutes/Clostridia | core set | chromosome | myDGR prediction |
| 148 | ADJS01006856.1 | uncultured Dorea sp. | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 149 | NZ_GG696046.1 | Ruminococcus sp. 5_1_39BFAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 150 | ADJS01008728.1 | uncultured Eubacterium sp. | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 151 | NZ_ACCL02000018.1 | Marvinbryantia formatexigens DSM 14469 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 152 | ADJT01012479.1 | uncultured Ruminococcus sp. | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 153 | ACTW01000035.1 | Lachnospiraceae bacterium 7_1_58FAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 154 | ACTW01000083.1 | Lachnospiraceae bacterium 7_1_58FAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 155 | NZ_AAYG02000002.1 | Ruminococcus gnavus ATCC 29149 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 156 | NZ_GL622409.1 | Lachnospiraceae bacterium 5_1_63FAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 157 | ADJT01005467.1 | uncultured Eubacterium sp. | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 158 | ABCB02000017.1 | Clostridium leptum DSM 753 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 159 | ADJS01009107.1 | uncultured Eubacterium sp. | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 160 | ACWK01000058.1 | Clostridium sp. 7_3_54FAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 161 | ACFX02000016.1 | Clostridium sp. M62/1 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 162 | JQ680361.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 163 | NZ_GL834357.1 | Clostridium symbiosum WAL-14673 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 164 | NZ_DS499683.1 | Clostridium scindens ATCC 35704 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 165 | NZ_JH114304.1 | Desulfovibrio sp. 6_1_46AFAA | Proteobacteria/Deltaproteobacteria | core set | WGS | myDGR prediction |
| 166 | NC_011420.2 | Rhodospirillum centenum SW | Proteobacteria/Alphaproteobacteria | core set | chromosome | myDGR prediction |
| 167 | AHMC01000020.1 | Desulfovibrio sp. U5L | Proteobacteria/Deltaproteobacteria | core set | WGS | myDGR prediction |
| 168 | AIDC01000066.1 | Vibrio cyclitrophicus 1F273 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 169 | NC_008750.1 | Shewanella sp. W3-18-1 | Proteobacteria/Gammaproteobacteria | core set | chromosome | myDGR prediction |
| 170 | NZ_ADOP01000022.1 | Pseudoalteromonas haloplanktis ANT/505 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 171 | FN297812.1 | Vibrio phage VP58.5 | Proteobacteria/Gammaproteobacteria | core set | phage | myDGR prediction |
| 172 | NC_004456.1 | Vibrio phage VHML | Proteobacteria/Gammaproteobacteria | core set | phage | myDGR prediction |
| 173 | AIDA01000015.1 | Vibrio cyclitrophicus 1F97 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 174 | NC_010113.1 | Vibrio sp. 0908 | Proteobacteria/Gammaproteobacteria | core set | plasmid | myDGR prediction |
| 175 | JQ680374.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 176 | ADDX01000030.1 | Ruminococcaceae bacterium D16 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 177 | NZ_JH376870.1 | Clostridium clostridioforme 2_1_49FAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 178 | NZ_GL945342.1 | Lachnospiraceae bacterium 2_1_58FAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 179 | NZ_GL945164.1 | Lachnospiraceae bacterium 1_4_56FAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 180 | ADJS01008728.1 | uncultured Eubacterium sp. | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 181 | JQ680377.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 182 | BAAV01000002.1 | human gut metagenome | Unidentified | core set | WGS | myDGR prediction |
| 183 | ABYO01000214.1 | Anaerococcus lactolyticus ATCC 51172 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 184 | ACTW01000029.1 | Lachnospiraceae bacterium 7_1_58FAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 185 | FP929045.1 | Faecalibacterium prausnitzii L2-6 | Firmicutes/Clostridia | core set | chromosome | myDGR prediction |
| 186 | JQ680375.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 187 | NZ_JH590875.1 | Lachnospiraceae bacterium 7_1_58FAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 188 | AMFD01000008.1 | Lactococcus garvieae I113 | Firmicutes/Bacilli | core set | WGS | myDGR prediction |
| 189 | AFNE01000030.1 | Ruminococcus flavefaciens 17 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 190 | FP929062.1 | butyrate-producing bacterium SS3/4 | Unidentified | core set | chromosome | myDGR prediction |
| 191 | NZ_GG667666.1 | Clostridium hathewayi DSM 13479 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 192 | NZ_JH376429.1 | Clostridium citroniae WAL-17108 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 193 | NZ_GL890521.1 | Lachnospiraceae bacterium 3_1_46FAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 194 | ADJS01011624.1 | uncultured Faecalibacterium sp. | Firmicutes/Clostridia | core set | metagenome | myDGR prediction |
| 195 | AFDI01000006.1 | Gardnerella vaginalis 315-A | Actinobacteria/Actinobacteria | core set | WGS | myDGR prediction |
| 196 | NZ_DS995479.1 | Collinsella stercoris DSM 13279 | Actinobacteria/Actinobacteria | core set | WGS | myDGR prediction |
| 197 | NC_013204.1 | Eggerthella lenta DSM 2243 | Actinobacteria/Actinobacteria | core set | chromosome | myDGR prediction |
| 198 | NZ_AAVN02000006.1 | Collinsella aerofaciens ATCC 25986 | Actinobacteria/Actinobacteria | core set | WGS | myDGR prediction |
| 199 | ADJT01003871.1 | uncultured Collinsella sp. | Actinobacteria/Actinobacteria | core set | WGS | myDGR prediction |
| 200 | NZ_GG692710.1 | Collinsella intestinalis DSM 13280 | Actinobacteria/Actinobacteria | core set | WGS | myDGR prediction |
| 201 | NZ_GG692714.1 | Roseburia intestinalis L1-82 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 202 | ADJS01010211.1 | uncultured Faecalibacterium sp. | Firmicutes/Clostridia | core set | metagenome | myDGR prediction |
| 203 | ACTW01000031.1 | Lachnospiraceae bacterium 7_1_58FAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 204 | ACTW01000029.1 | Lachnospiraceae bacterium 7_1_58FAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 205 | ADDX01000072.1 | Ruminococcaceae bacterium D16 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 206 | JQ680352.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 207 | JQ680373.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 208 | ADGC01006584.1 | wallaby gut metagenome | Unidentified | core set | metagenome | myDGR prediction |
| 209 | NC_009253.1 | Desulfotomaculum reducens MI-1 | Firmicutes/Clostridia | core set | chromosome | myDGR prediction |
| 210 | AMQJ01003071.1 | sediment metagenome | Unidentified | core set | metagenome | myDGR prediction |
| 211 | AFOP01000002.1 | Vibrio cholerae HE-09 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 212 | NZ_AKIY01000214.1 | Bradyrhizobium sp. YR681 | Proteobacteria/Alphaproteobacteria | core set | WGS | myDGR prediction |
| 213 | NC_005357.1 | Bordetella phage BPP-1 | Proteobacteria/Betaproteobacteria | core set | phage | myDGR prediction |
| 214 | NC_009428.1 | Rhodobacter sphaeroides ATCC 17025 | Proteobacteria/Alphaproteobacteria | core set | chromosome | myDGR prediction |
| 215 | NZ_GL949760.1 | Halomonas sp. TD01 | Proteobacteria/Gammaproteobacteria | core set | WGS | myDGR prediction |
| 216 | JQ680353.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 217 | NZ_JH376517.1 | Clostridium sp. 7_3_54FAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 218 | JQ680350.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 219 | JQ680358.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 220 | NZ_DS480690.1 | Clostridium bolteae ATCC BAA-613 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 221 | NZ_DS499697.1 | Clostridium scindens ATCC 35704 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 222 | CALI01000035.1 | Clostridiales bacterium 9401234 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 223 | NZ_GL982998.1 | Sporosarcina newyorkensis 2681 | Firmicutes/Bacilli | core set | WGS | myDGR prediction |
| 224 | NZ_AKKB01000035.1 | Brevibacillus sp. CF112 | Firmicutes/Bacilli | core set | WGS | myDGR prediction |
| 225 | NZ_AKKB01000053.1 | Brevibacillus sp. CF112 | Firmicutes/Bacilli | core set | WGS | myDGR prediction |
| 226 | ACIH01000097.1 | Paenibacillus sp. oral taxon 786 str. D14 | Firmicutes/Bacilli | core set | WGS | myDGR prediction |
| 227 | NC_015172.1 | Syntrophobotulus glycolicus DSM 8271 | Firmicutes/Clostridia | core set | chromosome | myDGR prediction |
| 228 | JQ680365.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 229 | JQ680355.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 230 | NZ_GG697157.2 | Faecalibacterium prausnitzii A2-165 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 231 | NZ_JH590866.1 | Lachnospiraceae bacterium 7_1_58FAA | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 232 | AMQJ01003125.1 | sediment metagenome | Unidentified | core set | metagenome | myDGR prediction |
| 233 | NC_010524.1 | Leptothrix cholodnii SP-6 | Proteobacteria/Betaproteobacteria | core set | chromosome | myDGR prediction |
| 234 | NC_007204.1 | Psychrobacter arcticus 273-4 | Proteobacteria/Gammaproteobacteria | core set | chromosome | myDGR prediction |
| 235 | CP001339.1 | Thioalkalivibrio sulfidophilus HL-EbGr7 | Proteobacteria/Gammaproteobacteria | core set | chromosome | myDGR prediction |
| 236 | HE576794.1 | Megasphaera elsdenii DSM 20460 | Firmicutes/Negativicutes | core set | chromosome | myDGR prediction |
| 237 | NC_013216.1 | Desulfotomaculum acetoxidans DSM 771 | Firmicutes/Clostridia | core set | chromosome | myDGR prediction |
| 238 | NC_009633.1 | Alkaliphilus metalliredigens QYMF | Firmicutes/Clostridia | core set | chromosome | myDGR prediction |
| 239 | NZ_CAKP01000036.1 | Caloramator australicus RC3 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 240 | JQ680366.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 241 | JQ680360.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 242 | NZ_DS264311.1 | Ruminococcus obeum ATCC 29174 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 243 | ADLQ01000071.1 | Clostridium symbiosum WAL-14163 | Firmicutes/Clostridia | core set | WGS | myDGR prediction |
| 244 | FP929053.1 | Ruminococcus sp. SR1/5 | Firmicutes/Clostridia | core set | chromosome | myDGR prediction |
| 245 | JQ680370.1 | unidentified phage | Unidentified | core set | phage | myDGR prediction |
| 246 | NZ_BALG01000089.1 | Paenibacillus popilliae ATCC 14706 | Firmicutes/Bacilli | core set | WGS | myDGR prediction |
Putative DGR systems in CPR set
| Index | Accession Number | Organism | Phylum/Class | DGR set | Type | DGR |
|---|---|---|---|---|---|---|
| 1 | AQSE01000026.1 | Nanoarchaeota archaeon SCGC AAA011-K22 | Nanoarchaeota/- | CPR set | WGS | myDGR prediction |
| 2 | AQSE01000048.1 | Nanoarchaeota archaeon SCGC AAA011-K22 | Nanoarchaeota/- | CPR set | WGS | myDGR prediction |
| 3 | AQSE01000038.1 | Nanoarchaeota archaeon SCGC AAA011-K22 | Nanoarchaeota/- | CPR set | WGS | myDGR prediction |
| 4 | AQSE01000039.1 | Nanoarchaeota archaeon SCGC AAA011-K22 | Nanoarchaeota/- | CPR set | WGS | myDGR prediction |
| 5 | KP703175.1 | ANMV-1 virus | Unidentified | CPR set | phage | myDGR prediction |
| 6 | CP005957.1 | Candidatus Saccharimonas aalborgensis | Unidentified | CPR set | chromosome | myDGR prediction |
| 7 | AMFJ01002827.1 | uncultured bacterium | Unidentified | CPR set | GWM | myDGR prediction |
| 8 | AMFJ01005734.1 | uncultured bacterium | Unidentified | CPR set | GWM | myDGR prediction |
| 9 | AMFJ01005865.1 | uncultured bacterium | Unidentified | CPR set | GWM | myDGR prediction |
| 10 | AMFJ01011991.1 | uncultured bacterium | Unidentified | CPR set | GWM | myDGR prediction |
| 11 | CP010426.1 | archaeon GW2011_AR20 | Unidentified | CPR set | GWM | myDGR prediction |
| 12 | JWKW01000020.1 | archaeon GW2011_AR17 | Unidentified | CPR set | GWM | myDGR prediction |
| 13 | LBOJ01000006.1 | Parcubacteria group bacterium GW2011_GWC2_32_10 | Unidentified | CPR set | GWM | myDGR prediction |
| 14 | LBOJ01000014.1 | Parcubacteria group bacterium GW2011_GWC2_32_10 | Unidentified | CPR set | GWM | myDGR prediction |
| 15 | LBOJ01000019.1 | Parcubacteria group bacterium GW2011_GWC2_32_10 | Unidentified | CPR set | GWM | myDGR prediction |
| 16 | LBOP01000015.1 | Candidatus Peregrinibacteria bacterium GW2011_GWF2_33_10 | Candidatus Peregrinibacteria/- | CPR set | GWM | myDGR prediction |
| 17 | LBOS01000005.1 | Parcubacteria group bacterium GW2011_GWA2_33_14 | Unidentified | CPR set | GWM | myDGR prediction |
| 18 | LBPL01000001.1 | Parcubacteria group bacterium GW2011_GWC1_34_10 | Unidentified | CPR set | GWM | myDGR prediction |
| 19 | LBPL01000003.1 | Parcubacteria group bacterium GW2011_GWC1_34_10 | Unidentified | CPR set | GWM | myDGR prediction |
| 20 | LBPO01000002.1 | Candidatus Magasanikbacteria bacterium GW2011_GWC2_34_16 | Candidatus Magasanikbacteria/- | CPR set | GWM | myDGR prediction |
| 21 | LBPW01000029.1 | Candidatus Nomurabacteria bacterium GW2011_GWF2_35_12 | Candidatus Nomurabacteria/- | CPR set | GWM | myDGR prediction |
| 22 | LBQA01000003.1 | Candidatus Moranbacteria bacterium GW2011_GWE1_35_17 | Candidatus Moranbacteria/- | CPR set | GWM | myDGR prediction |
| 23 | LBQD01000003.1 | Candidatus Moranbacteria bacterium GW2011_GWE2_35_2- | Candidatus Moranbacteria/- | CPR set | GWM | myDGR prediction |
| 24 | LBQM01000001.1 | Candidatus Moranbacteria bacterium GW2011_GWF2_35_39 | Candidatus Moranbacteria/- | CPR set | GWM | myDGR prediction |
| 25 | LBQO01000039.1 | Parcubacteria group bacterium GW2011_GWB1_35_5 | Unidentified | CPR set | GWM | myDGR prediction |
| 26 | LBQP01000024.1 | Candidatus Levybacteria bacterium GW2011_GWB1_35_5 | Candidatus Levybacteria/- | CPR set | GWM | myDGR prediction |
| 27 | LBRC01000027.1 | Parcubacteria group bacterium GW2011_GWA2_36_10 | Unidentified | CPR set | GWM | myDGR prediction |
| 28 | LBRD01000004.1 | Parcubacteria group bacterium GW2011_GWC1_36_108 | Unidentified | CPR set | GWM | myDGR prediction |
| 29 | LBRT01000013.1 | Parcubacteria group bacterium GW2011_GWA2_36_24 | Unidentified | CPR set | GWM | myDGR prediction |
| 30 | LBSG01000027.1 | Candidatus Moranbacteria bacterium GW2011_GWE1_36_7 | Candidatus Moranbacteria/- | CPR set | GWM | myDGR prediction |
| 31 | LBSI01000002.1 | Candidatus Moranbacteria bacterium GW2011_GWF1_36_78 | Candidatus Moranbacteria/- | CPR set | GWM | myDGR prediction |
| 32 | LBSX01000014.1 | Candidatus Magasanikbacteria bacterium GW2011_GWC2_37_14 | Candidatus Magasanikbacteria/- | CPR set | GWM | myDGR prediction |
| 33 | LBSY01000018.1 | Candidatus Levybacteria bacterium GW2011_GWA1_37_16 | Candidatus Levybacteria/- | CPR set | GWM | myDGR prediction |
| 34 | LBSY01000044.1 | Candidatus Levybacteria bacterium GW2011_GWA1_37_16 | Candidatus Levybacteria/- | CPR set | GWM | myDGR prediction |
| 35 | LBTF01000004.1 | Candidatus Nomurabacteria bacterium GW2011_GWB1_37_5 | Candidatus Nomurabacteria/- | CPR set | GWM | myDGR prediction |
| 36 | LBUP01000005.1 | Candidatus Daviesbacteria bacterium GW2011_GWA2_38_24 | Candidatus Daviesbacteria/- | CPR set | GWM | myDGR prediction |
| 37 | LBUR01000007.1 | Parcubacteria group bacterium GW2011_GWA2_38_27 | Unidentified | CPR set | GWM | myDGR prediction |
| 38 | LBUU01000007.1 | Candidatus Falkowbacteria bacterium GW2011_GWE1_38_31 | Candidatus Falkowbacteria/- | CPR set | GWM | myDGR prediction |
| 39 | LBVD01000002.1 | Parcubacteria group bacterium GW2011_GWA1_38_7 | Unidentified | CPR set | GWM | myDGR prediction |
| 40 | LBVH01000036.1 | Candidatus Daviesbacteria bacterium GW2011_GWF2_38_7 | Candidatus Daviesbacteria/- | CPR set | GWM | myDGR prediction |
| 41 | LBVK01000014.1 | Parcubacteria group bacterium GW2011_GWB1_38_8 | Unidentified | CPR set | GWM | myDGR prediction |
| 42 | LBVX01000030.1 | Candidatus Levybacteria bacterium GW2011_GWA1_39_11 | Candidatus Levybacteria/- | CPR set | GWM | myDGR prediction |
| 43 | LBWC01000004.1 | Parcubacteria group bacterium GW2011_GWC1_39_12 | Unidentified | CPR set | GWM | myDGR prediction |
| 44 | LBWF01000003.1 | Candidatus Yanofskybacteria bacterium GW2011_GWA1_39_13 | Candidatus Yanofskybacteria/- | CPR set | GWM | myDGR prediction |
| 45 | LBWH01000004.1 | Parcubacteria group bacterium GW2011_GWF2_39_13b | Unidentified | CPR set | GWM | myDGR prediction |
| 46 | LBWL01000009.1 | Candidatus Yanofskybacteria bacterium GW2011_GWD1_39_16 | Candidatus Yanofskybacteria/- | CPR set | GWM | myDGR prediction |
| 47 | LBXA01000009.1 | Candidatus Moranbacteria bacterium GW2011_GWA2_39_41 | Candidatus Moranbacteria/- | CPR set | GWM | myDGR prediction |
| 48 | LBXC01000004.1 | Candidatus Uhrbacteria bacterium GW2011_GWE1_39_46 | Candidatus Uhrbacteria/- | CPR set | GWM | myDGR prediction |
| 49 | LBXL01000029.1 | Candidatus Woesebacteria bacterium GW2011_GWA1_39_8 | Candidatus Woesebacteria/- | CPR set | GWM | myDGR prediction |
| 50 | LBXL01000035.1 | Candidatus Woesebacteria bacterium GW2011_GWA1_39_8 | Candidatus Woesebacteria/- | CPR set | GWM | myDGR prediction |
| 51 | LBXP01000008.1 | Parcubacteria group bacterium GW2011_GWF2_39_8b | Unidentified | CPR set | GWM | myDGR prediction |
| 52 | LBYD01000002.1 | Candidatus Moranbacteria bacterium GW2011_GWC2_40_12 | Candidatus Moranbacteria/- | CPR set | GWM | myDGR prediction |
| 53 | LBYE01000042.1 | Parcubacteria group bacterium GW2011_GWC1_40_13 | Unidentified | CPR set | GWM | myDGR prediction |
| 54 | LBYL01000001.1 | Candidatus Levybacteria bacterium GW2011_GWC1_40_19 | Candidatus Levybacteria/- | CPR set | GWM | myDGR prediction |
| 55 | LBYZ01000002.1 | Parcubacteria group bacterium GW2011_GWA2_40_37 | Unidentified | CPR set | GWM | myDGR prediction |
| 56 | LBZV01000004.1 | Candidatus Curtissbacteria bacterium GW2011_GWA1_40_9 | Candidatus Curtissbacteria/- | CPR set | GWM | myDGR prediction |
| 57 | LBZO01000013.1 | Candidatus Woesebacteria bacterium GW2011_GWA2_40_7 | Candidatus Woesebacteria/- | CPR set | GWM | myDGR prediction |
| 58 | LCAV01000017.1 | Candidatus Magasanikbacteria bacterium GW2011_GWC2_41_17 | Candidatus Magasanikbacteria/- | CPR set | GWM | myDGR prediction |
| 59 | LCBC01000017.1 | Candidatus Curtissbacteria bacterium GW2011_GWA2_41_24 | Candidatus Curtissbacteria/- | CPR set | GWM | myDGR prediction |
| 60 | LCBN01000004.1 | Candidatus Daviesbacteria bacterium GW2011_GWB1_41_5 | Candidatus Daviesbacteria/- | CPR set | GWM | myDGR prediction |
| 61 | LCBQ01000036.1 | Candidatus Yanofskybacteria bacterium GW2011_GWA1_41_6 | Candidatus Yanofskybacteria/- | CPR set | GWM | myDGR prediction |
| 62 | LCCB01000054.1 | Microgenomates group bacterium GW2011_GWC1_41_8 | Unidentified | CPR set | GWM | myDGR prediction |
| 63 | LCCP01000016.1 | Parcubacteria group bacterium GW2011_GWC2_42_12 | Unidentified | CPR set | GWM | myDGR prediction |
| 64 | LCCQ01000001.1 | Parcubacteria group bacterium GW2011_GWC2_42_13 | Unidentified | CPR set | GWM | myDGR prediction |
| 65 | LCCW01000010.1 | Candidatus Kuenenbacteria bacterium GW2011_GWA2_42_15 | Candidatus Kuenenbacteria/- | CPR set | GWM | myDGR prediction |
| 66 | LCDB01000024.1 | Candidatus Azambacteria bacterium GW2011_GWB1_42_17 | Candidatus Azambacteria/- | CPR set | GWM | myDGR prediction |
| 67 | LCDF01000006.1 | Candidatus Giovannonibacteria bacterium GW2011_GWF2_42_19 | Candidatus Giovannonibacteria/- | CPR set | GWM | myDGR prediction |
| 68 | LCDH01000011.1 | Parcubacteria group bacterium GW2011_GWC1_42_21 | Unidentified | CPR set | GWM | myDGR prediction |
| 69 | LCDS01000041.1 | Candidatus Nomurabacteria bacterium GW2011_GWA2_42_41 | Candidatus Nomurabacteria/- | CPR set | GWM | myDGR prediction |
| 70 | LCEB01000007.1 | Candidatus Daviesbacteria bacterium GW2011_GWA1_42_6 | Candidatus Daviesbacteria/- | CPR set | GWM | myDGR prediction |
| 71 | LCEC01000049.1 | Parcubacteria group bacterium GW2011_GWB1_42_6 | Unidentified | CPR set | GWM | myDGR prediction |
| 72 | LCEI01000004.1 | Parcubacteria group bacterium GW2011_GWA1_42_7 | Unidentified | CPR set | GWM | myDGR prediction |
| 73 | LCEP01000014.1 | Parcubacteria group bacterium GW2011_GWA2_42_80 | Unidentified | CPR set | GWM | myDGR prediction |
| 74 | LCES01000001.1 | Parcubacteria group bacterium GW2011_GWB1_42_9 | Unidentified | CPR set | GWM | myDGR prediction |
| 75 | LCES01000003.1 | Parcubacteria group bacterium GW2011_GWB1_42_9 | Unidentified | CPR set | GWM | myDGR prediction |
| 76 | LCEZ01000045.1 | Parcubacteria group bacterium GW2011_GWD2_43_10 | Unidentified | CPR set | GWM | myDGR prediction |
| 77 | LCFI01000001.1 | Candidatus Woesebacteria bacterium GW2011_GWA1_43_12 | Candidatus Woesebacteria/- | CPR set | GWM | myDGR prediction |
| 78 | LCFO01000002.1 | Candidatus Giovannonibacteria bacterium GW2011_GWB1_43_13 | Candidatus Giovannonibacteria/- | CPR set | GWM | myDGR prediction |
| 79 | LCGK01000002.1 | Parcubacteria group bacterium GW2011_GWC1_43_30 | Unidentified | CPR set | GWM | myDGR prediction |
| 80 | LCGL01000001.1 | Candidatus Falkowbacteria bacterium GW2011_GWF2_43_32 | Candidatus Falkowbacteria/- | CPR set | GWM | myDGR prediction |
| 81 | LCGO01000005.1 | Parcubacteria group bacterium GW2011_GWB1_43_6 | Unidentified | CPR set | GWM | myDGR prediction |
| 82 | LCGO01000017.1 | Parcubacteria group bacterium GW2011_GWB1_43_6 | Unidentified | CPR set | GWM | myDGR prediction |
| 83 | LCGW01000009.1 | Parcubacteria group bacterium GW2011_GWE1_43_8 | Unidentified | CPR set | GWM | myDGR prediction |
| 84 | LCHC01000022.1 | Parcubacteria group bacterium GW2011_GWF1_43_9 | Unidentified | CPR set | GWM | myDGR prediction |
| 85 | LCHE01000003.1 | Candidatus Yanofskybacteria bacterium GW2011_GWA2_44_10 | Candidatus Yanofskybacteria/- | CPR set | GWM | myDGR prediction |
| 86 | LCHF01000020.1 | Parcubacteria group bacterium GW2011_GWC1_44_10 | Unidentified | CPR set | GWM | myDGR prediction |
| 87 | LCHJ01000007.1 | Candidatus Moranbacteria bacterium GW2011_GWF2_44_10 | Candidatus Moranbacteria/- | CPR set | GWM | myDGR prediction |
| 88 | LCHM01000084.1 | Candidatus Gottesmanbacteria bacterium GW2011_GWB1_44_11c | Candidatus Gottesmanbacteria/- | CPR set | GWM | myDGR prediction |
| 89 | LCHS01000040.1 | Parcubacteria group bacterium GW2011_GWA2_44_13 | Unidentified | CPR set | GWM | myDGR prediction |
| 90 | LCHU01000008.1 | Candidatus Giovannonibacteria bacterium GW2011_GWA2_44_13b | Candidatus Giovannonibacteria/- | CPR set | GWM | myDGR prediction |
| 91 | LCIN01000007.1 | Candidatus Giovannonibacteria bacterium GW2011_GWB1_44_23 | Candidatus Giovannonibacteria/- | CPR set | GWM | myDGR prediction |
| 92 | LCJR01000002.1 | Candidatus Yanofskybacteria bacterium GW2011_GWA2_44_9 | Candidatus Yanofskybacteria/- | CPR set | GWM | myDGR prediction |
| 93 | LCKD01000002.1 | Candidatus Yanofskybacteria bacterium GW2011_GWB1_45_11 | Candidatus Yanofskybacteria/- | CPR set | GWM | myDGR prediction |
| 94 | LCKI01000003.1 | Parcubacteria group bacterium GW2011_GWC1_45_14 | Unidentified | CPR set | GWM | myDGR prediction |
| 95 | LCKK01000004.1 | Parcubacteria group bacterium GW2011_GWA2_45_15 | Unidentified | CPR set | GWM | myDGR prediction |
| 96 | LCLC01000018.1 | Parcubacteria group bacterium GW2011_GWA1_45_7 | Unidentified | CPR set | GWM | myDGR prediction |
| 97 | LCLM01000019.1 | Candidatus Woesebacteria bacterium GW2011_GWC2_45_9 | Candidatus Woesebacteria/- | CPR set | GWM | myDGR prediction |
| 98 | LCLQ01000001.1 | Parcubacteria group bacterium GW2011_GWA2_46_10 | Unidentified | CPR set | GWM | myDGR prediction |
| 99 | LCMF01000033.1 | candidate division WWE3 bacterium GW2011_GWA1_46_21 | candidate division WWE3/- | CPR set | GWM | myDGR prediction |
| 100 | LCMK01000005.1 | Candidatus Wolfebacteria bacterium GW2011_GWC2_46_275 | Candidatus Wolfebacteria/- | CPR set | GWM | myDGR prediction |
| 101 | LCML01000001.1 | Candidatus Beckwithbacteria bacterium GW2011_GWA1_46_30 | Candidatus Beckwithbacteria/- | CPR set | GWM | myDGR prediction |
| 102 | LCNF01000002.1 | Parcubacteria group bacterium GW2011_GWA1_47_10 | Unidentified | CPR set | GWM | myDGR prediction |
| 103 | LCNL01000022.1 | Parcubacteria group bacterium GW2011_GWA1_47_11 | Unidentified | CPR set | GWM | myDGR prediction |
| 104 | LCOG01000012.1 | Parcubacteria group bacterium GW2011_GWA2_47_26 | Unidentified | CPR set | GWM | myDGR prediction |
| 105 | LCOL01000016.1 | Parcubacteria group bacterium GW2011_GWA2_47_64 | Unidentified | CPR set | GWM | myDGR prediction |
| 106 | LCOP01000017.1 | Parcubacteria group bacterium GW2011_GWA1_47_8 | Unidentified | CPR set | GWM | myDGR prediction |
| 107 | LCOR01000029.1 | Parcubacteria group bacterium GW2011_GWA2_47_8 | Unidentified | CPR set | GWM | myDGR prediction |
| 108 | LCPI01000003.1 | Candidatus Jorgensenbacteria bacterium GW2011_GWA1_48_13 | Candidatus Jorgensenbacteria/- | CPR set | GWM | myDGR prediction |
| 109 | LCPM01000004.1 | Parcubacteria group bacterium GW2011_GWB1_48_6 | Unidentified | CPR set | GWM | myDGR prediction |
| 110 | LCQC01000006.1 | Parcubacteria group bacterium GW2011_GWB1_49_7 | Unidentified | CPR set | GWM | myDGR prediction |
| 111 | LCQJ01000030.1 | Parcubacteria group bacterium GW2011_GWA2_50_10b | Unidentified | CPR set | GWM | myDGR prediction |
| 112 | LCQW01000004.1 | Candidatus Kaiserbacteria bacterium GW2011_GWA2_52_12 | Candidatus Kaiserbacteria/- | CPR set | GWM | myDGR prediction |
| 113 | LCQY01000001.1 | candidate division Kazan bacterium GW2011_GWC1_52_13 | candidate division Kazan-3B-28/- | CPR set | GWM | myDGR prediction |
| 114 | LCRC01000045.1 | Parcubacteria group bacterium GW2011_GWB1_52_7 | Unidentified | CPR set | GWM | myDGR prediction |
| 115 | LCRK01000003.1 | Parcubacteria group bacterium GW2011_GWA2_53_21 | Unidentified | CPR set | GWM | myDGR prediction |
| 116 | LCRK01000008.1 | Parcubacteria group bacterium GW2011_GWA2_53_21 | Unidentified | CPR set | GWM | myDGR prediction |
| 117 | LCRU01000019.1 | Parcubacteria group bacterium GW2011_GWA1_54_9 | Unidentified | CPR set | GWM | myDGR prediction |
| 118 | LCRW01000003.1 | Parcubacteria group bacterium GW2011_GWB1_55_9 | Unidentified | CPR set | GWM | myDGR prediction |
| 119 | LCSB01000002.1 | Parcubacteria group bacterium GW2011_GWB1_56_8 | Unidentified | CPR set | GWM | myDGR prediction |
| 120 | LFCO01000003.1 | Parcubacteria bacterium C7867-005 | Candidatus Parcubacteria/- | CPR set | GWM | myDGR prediction |
Putative DGR systems in human microbiome dataset
| Index | Accession Number | Organism | DGR |
|---|---|---|---|
| 1 | SRS078176_LANL_scaffold_4398 | Human Metagenome | myDGR prediction |
| 2 | SRS078176_LANL_scaffold_28758 | Human Metagenome | myDGR prediction |
| 3 | SRS077730_LANL_scaffold_668 | Human Metagenome | myDGR prediction |
| 4 | SRS077730_LANL_scaffold_4938 | Human Metagenome | myDGR prediction |
| 5 | SRS077730_LANL_scaffold_24820 | Human Metagenome | myDGR prediction |
| 6 | SRS077730_LANL_scaffold_16194 | Human Metagenome | myDGR prediction |
| 7 | SRS077730_LANL_scaffold_13015 | Human Metagenome | myDGR prediction |
| 8 | SRS077730_LANL_scaffold_11133 | Human Metagenome | myDGR prediction |
| 9 | SRS065504_LANL_scaffold_46367 | Human Metagenome | myDGR prediction |
| 10 | SRS065504_LANL_scaffold_22142 | Human Metagenome | myDGR prediction |
| 11 | SRS065278_LANL_scaffold_62372 | Human Metagenome | myDGR prediction |
| 12 | SRS064774_LANL_scaffold_61312 | Human Metagenome | myDGR prediction |
| 13 | SRS064645_LANL_scaffold_3244 | Human Metagenome | myDGR prediction |
| 14 | SRS064557_LANL_scaffold_6895 | Human Metagenome | myDGR prediction |
| 15 | SRS064557_LANL_scaffold_20476 | Human Metagenome | myDGR prediction |
| 16 | SRS064557_LANL_scaffold_10761 | Human Metagenome | myDGR prediction |
| 17 | SRS064423_LANL_scaffold_46697 | Human Metagenome | myDGR prediction |
| 18 | SRS064276_LANL_scaffold_69655 | Human Metagenome | myDGR prediction |
| 19 | SRS064276_LANL_scaffold_69549 | Human Metagenome | myDGR prediction |
| 20 | SRS064276_LANL_scaffold_63194 | Human Metagenome | myDGR prediction |
| 21 | SRS064276_LANL_scaffold_37557 | Human Metagenome | myDGR prediction |
| 22 | SRS064276_LANL_scaffold_36918 | Human Metagenome | myDGR prediction |
| 23 | SRS064276_LANL_scaffold_14449 | Human Metagenome | myDGR prediction |
| 24 | SRS064276_LANL_scaffold_13002 | Human Metagenome | myDGR prediction |
| 25 | SRS064276_LANL_scaffold_12382 | Human Metagenome | myDGR prediction |
| 26 | SRS063985_LANL_scaffold_8871 | Human Metagenome | myDGR prediction |
| 27 | SRS063985_LANL_scaffold_6656 | Human Metagenome | myDGR prediction |
| 28 | SRS063985_LANL_scaffold_57289 | Human Metagenome | myDGR prediction |
| 29 | SRS063985_LANL_scaffold_50960 | Human Metagenome | myDGR prediction |
| 30 | SRS063985_LANL_scaffold_35341 | Human Metagenome | myDGR prediction |
| 31 | SRS063985_LANL_scaffold_33169 | Human Metagenome | myDGR prediction |
| 32 | SRS063985_LANL_scaffold_24478 | Human Metagenome | myDGR prediction |
| 33 | SRS063985_LANL_scaffold_23055 | Human Metagenome | myDGR prediction |
| 34 | SRS063985_LANL_scaffold_16562 | Human Metagenome | myDGR prediction |
| 35 | SRS063040_Baylor_scaffold_51362 | Human Metagenome | myDGR prediction |
| 36 | SRS063040_Baylor_scaffold_36078 | Human Metagenome | myDGR prediction |
| 37 | SRS063040_Baylor_scaffold_2907 | Human Metagenome | myDGR prediction |
| 38 | SRS062427_LANL_scaffold_6601 | Human Metagenome | myDGR prediction |
| 39 | SRS062427_LANL_scaffold_13273 | Human Metagenome | myDGR prediction |
| 40 | SRS062427_LANL_scaffold_11801 | Human Metagenome | myDGR prediction |
| 41 | SRS058770_LANL_scaffold_4025 | Human Metagenome | myDGR prediction |
| 42 | SRS058770_LANL_scaffold_39374 | Human Metagenome | myDGR prediction |
| 43 | SRS058770_LANL_scaffold_35718 | Human Metagenome | myDGR prediction |
| 44 | SRS058770_LANL_scaffold_26875 | Human Metagenome | myDGR prediction |
| 45 | SRS058770_LANL_scaffold_24560 | Human Metagenome | myDGR prediction |
| 46 | SRS058723_Baylor_scaffold_5897 | Human Metagenome | myDGR prediction |
| 47 | SRS058723_Baylor_scaffold_12543 | Human Metagenome | myDGR prediction |
| 48 | SRS058723_Baylor_scaffold_1072 | Human Metagenome | myDGR prediction |
| 49 | SRS057791_LANL_scaffold_13865 | Human Metagenome | myDGR prediction |
| 50 | SRS057717_LANL_scaffold_6837 | Human Metagenome | myDGR prediction |
| 51 | SRS057717_LANL_scaffold_2749 | Human Metagenome | myDGR prediction |
| 52 | SRS057539_LANL_scaffold_61511 | Human Metagenome | myDGR prediction |
| 53 | SRS056622_LANL_scaffold_37409 | Human Metagenome | myDGR prediction |
| 54 | SRS056519_LANL_scaffold_25052 | Human Metagenome | myDGR prediction |
| 55 | SRS056519_LANL_scaffold_2340 | Human Metagenome | myDGR prediction |
| 56 | SRS056519_LANL_scaffold_19792 | Human Metagenome | myDGR prediction |
| 57 | SRS056259_LANL_scaffold_58167 | Human Metagenome | myDGR prediction |
| 58 | SRS056259_LANL_scaffold_48800 | Human Metagenome | myDGR prediction |
| 59 | SRS056259_LANL_scaffold_17268 | Human Metagenome | myDGR prediction |
| 60 | SRS056259_LANL_scaffold_15498 | Human Metagenome | myDGR prediction |
| 61 | SRS056259_LANL_scaffold_10150 | Human Metagenome | myDGR prediction |
| 62 | SRS055982_Baylor_scaffold_6895 | Human Metagenome | myDGR prediction |
| 63 | SRS055982_Baylor_scaffold_5011 | Human Metagenome | myDGR prediction |
| 64 | SRS055982_Baylor_scaffold_3547 | Human Metagenome | myDGR prediction |
| 65 | SRS055982_Baylor_scaffold_3508 | Human Metagenome | myDGR prediction |
| 66 | SRS055982_Baylor_scaffold_18529 | Human Metagenome | myDGR prediction |
| 67 | SRS055982_Baylor_scaffold_18092 | Human Metagenome | myDGR prediction |
| 68 | SRS055982_Baylor_scaffold_17347 | Human Metagenome | myDGR prediction |
| 69 | SRS054956_LANL_scaffold_45718 | Human Metagenome | myDGR prediction |
| 70 | SRS054956_LANL_scaffold_45565 | Human Metagenome | myDGR prediction |
| 71 | SRS054956_LANL_scaffold_43679 | Human Metagenome | myDGR prediction |
| 72 | SRS054956_LANL_scaffold_38823 | Human Metagenome | myDGR prediction |
| 73 | SRS054956_LANL_scaffold_15399 | Human Metagenome | myDGR prediction |
| 74 | SRS054956_LANL_scaffold_12271 | Human Metagenome | myDGR prediction |
| 75 | SRS054590_LANL_scaffold_823 | Human Metagenome | myDGR prediction |
| 76 | SRS054590_LANL_scaffold_2693 | Human Metagenome | myDGR prediction |
| 77 | SRS054590_LANL_scaffold_1726 | Human Metagenome | myDGR prediction |
| 78 | SRS053603_LANL_scaffold_49461 | Human Metagenome | myDGR prediction |
| 79 | SRS053398_LANL_scaffold_6921 | Human Metagenome | myDGR prediction |
| 80 | SRS053398_LANL_scaffold_3283 | Human Metagenome | myDGR prediction |
| 81 | SRS053398_LANL_scaffold_24779 | Human Metagenome | myDGR prediction |
| 82 | SRS053335_LANL_scaffold_26347 | Human Metagenome | myDGR prediction |
| 83 | SRS053335_LANL_scaffold_12895 | Human Metagenome | myDGR prediction |
| 84 | SRS052697_LANL_scaffold_75585 | Human Metagenome | myDGR prediction |
| 85 | SRS052697_LANL_scaffold_63124 | Human Metagenome | myDGR prediction |
| 86 | SRS052697_LANL_scaffold_46101 | Human Metagenome | myDGR prediction |
| 87 | SRS052697_LANL_scaffold_26371 | Human Metagenome | myDGR prediction |
| 88 | SRS052697_LANL_scaffold_15219 | Human Metagenome | myDGR prediction |
| 89 | SRS052697_LANL_scaffold_12111 | Human Metagenome | myDGR prediction |
| 90 | SRS052027_LANL_scaffold_64 | Human Metagenome | myDGR prediction |
| 91 | SRS052027_LANL_scaffold_4315 | Human Metagenome | myDGR prediction |
| 92 | SRS052027_LANL_scaffold_25763 | Human Metagenome | myDGR prediction |
| 93 | SRS052027_LANL_scaffold_25529 | Human Metagenome | myDGR prediction |
| 94 | SRS052027_LANL_scaffold_24753 | Human Metagenome | myDGR prediction |
| 95 | SRS052027_LANL_scaffold_24545 | Human Metagenome | myDGR prediction |
| 96 | SRS052027_LANL_scaffold_24280 | Human Metagenome | myDGR prediction |
| 97 | SRS052027_LANL_scaffold_10277 | Human Metagenome | myDGR prediction |
| 98 | SRS051882_Baylor_scaffold_7329 | Human Metagenome | myDGR prediction |
| 99 | SRS051882_Baylor_scaffold_1184 | Human Metagenome | myDGR prediction |
| 100 | SRS051031_LANL_scaffold_8810 | Human Metagenome | myDGR prediction |
| 101 | SRS051031_LANL_scaffold_65435 | Human Metagenome | myDGR prediction |
| 102 | SRS051031_LANL_scaffold_62496 | Human Metagenome | myDGR prediction |
| 103 | SRS051031_LANL_scaffold_4812 | Human Metagenome | myDGR prediction |
| 104 | SRS051031_LANL_scaffold_22143 | Human Metagenome | myDGR prediction |
| 105 | SRS051031_LANL_scaffold_12877 | Human Metagenome | myDGR prediction |
| 106 | SRS050925_LANL_scaffold_74183 | Human Metagenome | myDGR prediction |
| 107 | SRS050925_LANL_scaffold_73778 | Human Metagenome | myDGR prediction |
| 108 | SRS050925_LANL_scaffold_73619 | Human Metagenome | myDGR prediction |
| 109 | SRS050925_LANL_scaffold_63133 | Human Metagenome | myDGR prediction |
| 110 | SRS050925_LANL_scaffold_59006 | Human Metagenome | myDGR prediction |
| 111 | SRS050925_LANL_scaffold_57202 | Human Metagenome | myDGR prediction |
| 112 | SRS050925_LANL_scaffold_55373 | Human Metagenome | myDGR prediction |
| 113 | SRS050925_LANL_scaffold_5503 | Human Metagenome | myDGR prediction |
| 114 | SRS050925_LANL_scaffold_48374 | Human Metagenome | myDGR prediction |
| 115 | SRS050925_LANL_scaffold_32081 | Human Metagenome | myDGR prediction |
| 116 | SRS050925_LANL_scaffold_23304 | Human Metagenome | myDGR prediction |
| 117 | SRS050925_LANL_scaffold_21627 | Human Metagenome | myDGR prediction |
| 118 | SRS050925_LANL_scaffold_16337 | Human Metagenome | myDGR prediction |
| 119 | SRS050925_LANL_scaffold_11621 | Human Metagenome | myDGR prediction |
| 120 | SRS050925_LANL_scaffold_10132 | Human Metagenome | myDGR prediction |
| 121 | SRS050752_LANL_scaffold_9299 | Human Metagenome | myDGR prediction |
| 122 | SRS050752_LANL_scaffold_29730 | Human Metagenome | myDGR prediction |
| 123 | SRS050752_LANL_scaffold_25295 | Human Metagenome | myDGR prediction |
| 124 | SRS050752_LANL_scaffold_24509 | Human Metagenome | myDGR prediction |
| 125 | SRS050752_LANL_scaffold_21568 | Human Metagenome | myDGR prediction |
| 126 | SRS050422_LANL_scaffold_95410 | Human Metagenome | myDGR prediction |
| 127 | SRS050422_LANL_scaffold_82453 | Human Metagenome | myDGR prediction |
| 128 | SRS050422_LANL_scaffold_50 | Human Metagenome | myDGR prediction |
| 129 | SRS050422_LANL_scaffold_49109 | Human Metagenome | myDGR prediction |
| 130 | SRS050422_LANL_scaffold_42845 | Human Metagenome | myDGR prediction |
| 131 | SRS050422_LANL_scaffold_32447 | Human Metagenome | myDGR prediction |
| 132 | SRS050299_LANL_scaffold_687 | Human Metagenome | myDGR prediction |
| 133 | SRS050299_LANL_scaffold_5926 | Human Metagenome | myDGR prediction |
| 134 | SRS050299_LANL_scaffold_30827 | Human Metagenome | myDGR prediction |
| 135 | SRS050299_LANL_scaffold_23554 | Human Metagenome | myDGR prediction |
| 136 | SRS050299_LANL_scaffold_23470 | Human Metagenome | myDGR prediction |
| 137 | SRS050299_LANL_scaffold_14550 | Human Metagenome | myDGR prediction |
| 138 | SRS049995_LANL_scaffold_4188 | Human Metagenome | myDGR prediction |
| 139 | SRS049995_LANL_scaffold_40118 | Human Metagenome | myDGR prediction |
| 140 | SRS049995_LANL_scaffold_39136 | Human Metagenome | myDGR prediction |
| 141 | SRS049995_LANL_scaffold_38903 | Human Metagenome | myDGR prediction |
| 142 | SRS049995_LANL_scaffold_10062 | Human Metagenome | myDGR prediction |
| 143 | SRS049959_WUGC_scaffold_738 | Human Metagenome | myDGR prediction |
| 144 | SRS049959_WUGC_scaffold_51619 | Human Metagenome | myDGR prediction |
| 145 | SRS049959_WUGC_scaffold_42809 | Human Metagenome | myDGR prediction |
| 146 | SRS049959_WUGC_scaffold_31150 | Human Metagenome | myDGR prediction |
| 147 | SRS049959_WUGC_scaffold_2163 | Human Metagenome | myDGR prediction |
| 148 | SRS048870_WUGC_scaffold_8930 | Human Metagenome | myDGR prediction |
| 149 | SRS048870_WUGC_scaffold_49996 | Human Metagenome | myDGR prediction |
| 150 | SRS048870_WUGC_scaffold_49190 | Human Metagenome | myDGR prediction |
| 151 | SRS048870_WUGC_scaffold_37391 | Human Metagenome | myDGR prediction |
| 152 | SRS048870_WUGC_scaffold_14906 | Human Metagenome | myDGR prediction |
| 153 | SRS048870_WUGC_scaffold_13953 | Human Metagenome | myDGR prediction |
| 154 | SRS048870_WUGC_scaffold_13545 | Human Metagenome | myDGR prediction |
| 155 | SRS048164_WUGC_scaffold_8644 | Human Metagenome | myDGR prediction |
| 156 | SRS048164_WUGC_scaffold_7733 | Human Metagenome | myDGR prediction |
| 157 | SRS048164_WUGC_scaffold_29975 | Human Metagenome | myDGR prediction |
| 158 | SRS048164_WUGC_scaffold_28697 | Human Metagenome | myDGR prediction |
| 159 | SRS048164_WUGC_scaffold_24970 | Human Metagenome | myDGR prediction |
| 160 | SRS048164_WUGC_scaffold_16288 | Human Metagenome | myDGR prediction |
| 161 | SRS047044_WUGC_scaffold_7491 | Human Metagenome | myDGR prediction |
| 162 | SRS047044_WUGC_scaffold_30551 | Human Metagenome | myDGR prediction |
| 163 | SRS047044_WUGC_scaffold_29602 | Human Metagenome | myDGR prediction |
| 164 | SRS047044_WUGC_scaffold_29536 | Human Metagenome | myDGR prediction |
| 165 | SRS047044_WUGC_scaffold_28202 | Human Metagenome | myDGR prediction |
| 166 | SRS047044_WUGC_scaffold_23650 | Human Metagenome | myDGR prediction |
| 167 | SRS047044_WUGC_scaffold_1816 | Human Metagenome | myDGR prediction |
| 168 | SRS047014_WUGC_scaffold_80783 | Human Metagenome | myDGR prediction |
| 169 | SRS047014_WUGC_scaffold_73533 | Human Metagenome | myDGR prediction |
| 170 | SRS047014_WUGC_scaffold_64822 | Human Metagenome | myDGR prediction |
| 171 | SRS047014_WUGC_scaffold_12040 | Human Metagenome | myDGR prediction |
| 172 | SRS045715_LANL_scaffold_24840 | Human Metagenome | myDGR prediction |
| 173 | SRS045713_WUGC_scaffold_17477 | Human Metagenome | myDGR prediction |
| 174 | SRS045645_Baylor_scaffold_11758 | Human Metagenome | myDGR prediction |
| 175 | SRS044373_WUGC_scaffold_57191 | Human Metagenome | myDGR prediction |
| 176 | SRS044373_WUGC_scaffold_24244 | Human Metagenome | myDGR prediction |
| 177 | SRS043701_LANL_scaffold_6522 | Human Metagenome | myDGR prediction |
| 178 | SRS043701_LANL_scaffold_4863 | Human Metagenome | myDGR prediction |
| 179 | SRS043701_LANL_scaffold_31205 | Human Metagenome | myDGR prediction |
| 180 | SRS043701_LANL_scaffold_20843 | Human Metagenome | myDGR prediction |
| 181 | SRS043411_WUGC_scaffold_42074 | Human Metagenome | myDGR prediction |
| 182 | SRS043411_WUGC_scaffold_28831 | Human Metagenome | myDGR prediction |
| 183 | SRS043411_WUGC_scaffold_28490 | Human Metagenome | myDGR prediction |
| 184 | SRS043411_WUGC_scaffold_21308 | Human Metagenome | myDGR prediction |
| 185 | SRS043411_WUGC_scaffold_18861 | Human Metagenome | myDGR prediction |
| 186 | SRS043411_WUGC_scaffold_1881 | Human Metagenome | myDGR prediction |
| 187 | SRS043411_WUGC_scaffold_10108 | Human Metagenome | myDGR prediction |
| 188 | SRS043001_WUGC_scaffold_43290 | Human Metagenome | myDGR prediction |
| 189 | SRS042643_WUGC_scaffold_19693 | Human Metagenome | myDGR prediction |
| 190 | SRS042628_WUGC_scaffold_29665 | Human Metagenome | myDGR prediction |
| 191 | SRS042628_WUGC_scaffold_2354 | Human Metagenome | myDGR prediction |
| 192 | SRS042284_WUGC_scaffold_8810 | Human Metagenome | myDGR prediction |
| 193 | SRS042284_WUGC_scaffold_4374 | Human Metagenome | myDGR prediction |
| 194 | SRS042131_WUGC_scaffold_61116 | Human Metagenome | myDGR prediction |
| 195 | SRS024625_LANL_scaffold_16620 | Human Metagenome | myDGR prediction |
| 196 | SRS024549_LANL_scaffold_4925 | Human Metagenome | myDGR prediction |
| 197 | SRS024435_LANL_scaffold_33569 | Human Metagenome | myDGR prediction |
| 198 | SRS024435_LANL_scaffold_33181 | Human Metagenome | myDGR prediction |
| 199 | SRS024435_LANL_scaffold_2449 | Human Metagenome | myDGR prediction |
| 200 | SRS024435_LANL_scaffold_1610 | Human Metagenome | myDGR prediction |
| 201 | SRS024388_LANL_scaffold_15345 | Human Metagenome | myDGR prediction |
| 202 | SRS024331_LANL_scaffold_48022 | Human Metagenome | myDGR prediction |
| 203 | SRS024331_LANL_scaffold_47838 | Human Metagenome | myDGR prediction |
| 204 | SRS024331_LANL_scaffold_43108 | Human Metagenome | myDGR prediction |
| 205 | SRS024265_LANL_scaffold_6549 | Human Metagenome | myDGR prediction |
| 206 | SRS024265_LANL_scaffold_29936 | Human Metagenome | myDGR prediction |
| 207 | SRS024132_Baylor_scaffold_7719 | Human Metagenome | myDGR prediction |
| 208 | SRS024132_Baylor_scaffold_34478 | Human Metagenome | myDGR prediction |
| 209 | SRS024132_Baylor_scaffold_33615 | Human Metagenome | myDGR prediction |
| 210 | SRS024132_Baylor_scaffold_18469 | Human Metagenome | myDGR prediction |
| 211 | SRS024132_Baylor_scaffold_11143 | Human Metagenome | myDGR prediction |
| 212 | SRS024075_LANL_scaffold_3043 | Human Metagenome | myDGR prediction |
| 213 | SRS024075_LANL_scaffold_24173 | Human Metagenome | myDGR prediction |
| 214 | SRS024075_LANL_scaffold_22810 | Human Metagenome | myDGR prediction |
| 215 | SRS024075_LANL_scaffold_16737 | Human Metagenome | myDGR prediction |
| 216 | SRS024075_LANL_scaffold_1103 | Human Metagenome | myDGR prediction |
| 217 | SRS023914_Baylor_scaffold_8987 | Human Metagenome | myDGR prediction |
| 218 | SRS023914_Baylor_scaffold_5263 | Human Metagenome | myDGR prediction |
| 219 | SRS023914_Baylor_scaffold_4680 | Human Metagenome | myDGR prediction |
| 220 | SRS023914_Baylor_scaffold_3863 | Human Metagenome | myDGR prediction |
| 221 | SRS023914_Baylor_scaffold_35241 | Human Metagenome | myDGR prediction |
| 222 | SRS023914_Baylor_scaffold_34958 | Human Metagenome | myDGR prediction |
| 223 | SRS023914_Baylor_scaffold_30401 | Human Metagenome | myDGR prediction |
| 224 | SRS023914_Baylor_scaffold_20705 | Human Metagenome | myDGR prediction |
| 225 | SRS023914_Baylor_scaffold_11213 | Human Metagenome | myDGR prediction |
| 226 | SRS023583_Baylor_scaffold_6064 | Human Metagenome | myDGR prediction |
| 227 | SRS023526_Baylor_scaffold_6887 | Human Metagenome | myDGR prediction |
| 228 | SRS023526_Baylor_scaffold_35880 | Human Metagenome | myDGR prediction |
| 229 | SRS023526_Baylor_scaffold_30045 | Human Metagenome | myDGR prediction |
| 230 | SRS023352_LANL_scaffold_21098 | Human Metagenome | myDGR prediction |
| 231 | SRS023346_LANL_scaffold_1239 | Human Metagenome | myDGR prediction |
| 232 | SRS023176_Baylor_scaffold_2758 | Human Metagenome | myDGR prediction |
| 233 | SRS022609_Baylor_scaffold_90461 | Human Metagenome | myDGR prediction |
| 234 | SRS022609_Baylor_scaffold_84071 | Human Metagenome | myDGR prediction |
| 235 | SRS022609_Baylor_scaffold_56409 | Human Metagenome | myDGR prediction |
| 236 | SRS022609_Baylor_scaffold_55782 | Human Metagenome | myDGR prediction |
| 237 | SRS022609_Baylor_scaffold_37769 | Human Metagenome | myDGR prediction |
| 238 | SRS022609_Baylor_scaffold_27140 | Human Metagenome | myDGR prediction |
| 239 | SRS022609_Baylor_scaffold_19029 | Human Metagenome | myDGR prediction |
| 240 | SRS020869_Baylor_scaffold_979 | Human Metagenome | myDGR prediction |
| 241 | SRS020869_Baylor_scaffold_3601 | Human Metagenome | myDGR prediction |
| 242 | SRS020328_Baylor_scaffold_37240 | Human Metagenome | myDGR prediction |
| 243 | SRS020328_Baylor_scaffold_34917 | Human Metagenome | myDGR prediction |
| 244 | SRS020328_Baylor_scaffold_30660 | Human Metagenome | myDGR prediction |
| 245 | SRS020328_Baylor_scaffold_20831 | Human Metagenome | myDGR prediction |
| 246 | SRS019968_Baylor_scaffold_4282 | Human Metagenome | myDGR prediction |
| 247 | SRS019968_Baylor_scaffold_19401 | Human Metagenome | myDGR prediction |
| 248 | SRS019910_WUGC_scaffold_78514 | Human Metagenome | myDGR prediction |
| 249 | SRS019910_WUGC_scaffold_5688 | Human Metagenome | myDGR prediction |
| 250 | SRS019910_WUGC_scaffold_48843 | Human Metagenome | myDGR prediction |
| 251 | SRS019787_WUGC_scaffold_26812 | Human Metagenome | myDGR prediction |
| 252 | SRS019787_WUGC_scaffold_25006 | Human Metagenome | myDGR prediction |
| 253 | SRS019787_WUGC_scaffold_11166 | Human Metagenome | myDGR prediction |
| 254 | SRS019601_WUGC_scaffold_29116 | Human Metagenome | myDGR prediction |
| 255 | SRS019582_WUGC_scaffold_9518 | Human Metagenome | myDGR prediction |
| 256 | SRS019582_WUGC_scaffold_88355 | Human Metagenome | myDGR prediction |
| 257 | SRS019582_WUGC_scaffold_74205 | Human Metagenome | myDGR prediction |
| 258 | SRS019582_WUGC_scaffold_26869 | Human Metagenome | myDGR prediction |
| 259 | SRS019582_WUGC_scaffold_22380 | Human Metagenome | myDGR prediction |
| 260 | SRS019582_WUGC_scaffold_22156 | Human Metagenome | myDGR prediction |
| 261 | SRS019582_WUGC_scaffold_11793 | Human Metagenome | myDGR prediction |
| 262 | SRS019397_WUGC_scaffold_24605 | Human Metagenome | myDGR prediction |
| 263 | SRS019267_WUGC_scaffold_8107 | Human Metagenome | myDGR prediction |
| 264 | SRS019219_WUGC_scaffold_31979 | Human Metagenome | myDGR prediction |
| 265 | SRS019161_WUGC_scaffold_67361 | Human Metagenome | myDGR prediction |
| 266 | SRS019161_WUGC_scaffold_62155 | Human Metagenome | myDGR prediction |
| 267 | SRS019161_WUGC_scaffold_35145 | Human Metagenome | myDGR prediction |
| 268 | SRS019161_WUGC_scaffold_33958 | Human Metagenome | myDGR prediction |
| 269 | SRS019161_WUGC_scaffold_17514 | Human Metagenome | myDGR prediction |
| 270 | SRS019161_WUGC_scaffold_16553 | Human Metagenome | myDGR prediction |
| 271 | SRS019161_WUGC_scaffold_12222 | Human Metagenome | myDGR prediction |
| 272 | SRS019045_WUGC_scaffold_59458 | Human Metagenome | myDGR prediction |
| 273 | SRS019030_Baylor_scaffold_39776 | Human Metagenome | myDGR prediction |
| 274 | SRS019030_Baylor_scaffold_25174 | Human Metagenome | myDGR prediction |
| 275 | SRS019030_Baylor_scaffold_15832 | Human Metagenome | myDGR prediction |
| 276 | SRS018969_WUGC_scaffold_17555 | Human Metagenome | myDGR prediction |
| 277 | SRS018817_WUGC_scaffold_4993 | Human Metagenome | myDGR prediction |
| 278 | SRS018817_WUGC_scaffold_24201 | Human Metagenome | myDGR prediction |
| 279 | SRS018817_WUGC_scaffold_11409 | Human Metagenome | myDGR prediction |
| 280 | SRS018739_WUGC_scaffold_59882 | Human Metagenome | myDGR prediction |
| 281 | SRS018656_WUGC_scaffold_9190 | Human Metagenome | myDGR prediction |
| 282 | SRS018656_WUGC_scaffold_1644 | Human Metagenome | myDGR prediction |
| 283 | SRS018575_WUGC_scaffold_9460 | Human Metagenome | myDGR prediction |
| 284 | SRS018427_Baylor_scaffold_572 | Human Metagenome | myDGR prediction |
| 285 | SRS018351_Baylor_scaffold_29504 | Human Metagenome | myDGR prediction |
| 286 | SRS018351_Baylor_scaffold_19307 | Human Metagenome | myDGR prediction |
| 287 | SRS018351_Baylor_scaffold_12783 | Human Metagenome | myDGR prediction |
| 288 | SRS017821_Baylor_scaffold_7166 | Human Metagenome | myDGR prediction |
| 289 | SRS017821_Baylor_scaffold_5879 | Human Metagenome | myDGR prediction |
| 290 | SRS017821_Baylor_scaffold_547 | Human Metagenome | myDGR prediction |
| 291 | SRS017821_Baylor_scaffold_3688 | Human Metagenome | myDGR prediction |
| 292 | SRS017821_Baylor_scaffold_3398 | Human Metagenome | myDGR prediction |
| 293 | SRS017821_Baylor_scaffold_20533 | Human Metagenome | myDGR prediction |
| 294 | SRS017821_Baylor_scaffold_19389 | Human Metagenome | myDGR prediction |
| 295 | SRS017808_Baylor_scaffold_7236 | Human Metagenome | myDGR prediction |
| 296 | SRS017701_Baylor_scaffold_17293 | Human Metagenome | myDGR prediction |
| 297 | SRS017701_Baylor_scaffold_12533 | Human Metagenome | myDGR prediction |
| 298 | SRS017701_Baylor_scaffold_1224 | Human Metagenome | myDGR prediction |
| 299 | SRS017701_Baylor_scaffold_11281 | Human Metagenome | myDGR prediction |
| 300 | SRS017521_Baylor_scaffold_6683 | Human Metagenome | myDGR prediction |
| 301 | SRS017521_Baylor_scaffold_45782 | Human Metagenome | myDGR prediction |
| 302 | SRS017521_Baylor_scaffold_30298 | Human Metagenome | myDGR prediction |
| 303 | SRS017521_Baylor_scaffold_17890 | Human Metagenome | myDGR prediction |
| 304 | SRS017433_Baylor_scaffold_19382 | Human Metagenome | myDGR prediction |
| 305 | SRS017307_Baylor_scaffold_9532 | Human Metagenome | myDGR prediction |
| 306 | SRS017307_Baylor_scaffold_8450 | Human Metagenome | myDGR prediction |
| 307 | SRS017307_Baylor_scaffold_4293 | Human Metagenome | myDGR prediction |
| 308 | SRS017307_Baylor_scaffold_33015 | Human Metagenome | myDGR prediction |
| 309 | SRS017307_Baylor_scaffold_32950 | Human Metagenome | myDGR prediction |
| 310 | SRS017307_Baylor_scaffold_30143 | Human Metagenome | myDGR prediction |
| 311 | SRS017307_Baylor_scaffold_25643 | Human Metagenome | myDGR prediction |
| 312 | SRS017247_LANL_scaffold_10233 | Human Metagenome | myDGR prediction |
| 313 | SRS017191_Baylor_scaffold_3420 | Human Metagenome | myDGR prediction |
| 314 | SRS016989_Baylor_scaffold_55 | Human Metagenome | myDGR prediction |
| 315 | SRS016569_Baylor_scaffold_10980 | Human Metagenome | myDGR prediction |
| 316 | SRS016335_WUGC_scaffold_5786 | Human Metagenome | myDGR prediction |
| 317 | SRS016335_WUGC_scaffold_42665 | Human Metagenome | myDGR prediction |
| 318 | SRS016335_WUGC_scaffold_42155 | Human Metagenome | myDGR prediction |
| 319 | SRS016335_WUGC_scaffold_32876 | Human Metagenome | myDGR prediction |
| 320 | SRS016335_WUGC_scaffold_11952 | Human Metagenome | myDGR prediction |
| 321 | SRS016203_WUGC_scaffold_853 | Human Metagenome | myDGR prediction |
| 322 | SRS016203_WUGC_scaffold_4021 | Human Metagenome | myDGR prediction |
| 323 | SRS016095_WUGC_scaffold_7254 | Human Metagenome | myDGR prediction |
| 324 | SRS016095_WUGC_scaffold_2324 | Human Metagenome | myDGR prediction |
| 325 | SRS016095_WUGC_scaffold_20077 | Human Metagenome | myDGR prediction |
| 326 | SRS016095_WUGC_scaffold_16169 | Human Metagenome | myDGR prediction |
| 327 | SRS016095_WUGC_scaffold_15302 | Human Metagenome | myDGR prediction |
| 328 | SRS016086_WUGC_scaffold_26885 | Human Metagenome | myDGR prediction |
| 329 | SRS016056_WUGC_scaffold_8658 | Human Metagenome | myDGR prediction |
| 330 | SRS016056_WUGC_scaffold_1971 | Human Metagenome | myDGR prediction |
| 331 | SRS016056_WUGC_scaffold_1338 | Human Metagenome | myDGR prediction |
| 332 | SRS016018_WUGC_scaffold_9927 | Human Metagenome | myDGR prediction |
| 333 | SRS016018_WUGC_scaffold_5565 | Human Metagenome | myDGR prediction |
| 334 | SRS016018_WUGC_scaffold_4418 | Human Metagenome | myDGR prediction |
| 335 | SRS016018_WUGC_scaffold_4060 | Human Metagenome | myDGR prediction |
| 336 | SRS016018_WUGC_scaffold_22555 | Human Metagenome | myDGR prediction |
| 337 | SRS016018_WUGC_scaffold_20700 | Human Metagenome | myDGR prediction |
| 338 | SRS016002_WUGC_scaffold_62909 | Human Metagenome | myDGR prediction |
| 339 | SRS016002_WUGC_scaffold_62244 | Human Metagenome | myDGR prediction |
| 340 | SRS015960_WUGC_scaffold_9632 | Human Metagenome | myDGR prediction |
| 341 | SRS015960_WUGC_scaffold_32057 | Human Metagenome | myDGR prediction |
| 342 | SRS015960_WUGC_scaffold_31981 | Human Metagenome | myDGR prediction |
| 343 | SRS015960_WUGC_scaffold_27365 | Human Metagenome | myDGR prediction |
| 344 | SRS015941_WUGC_scaffold_64627 | Human Metagenome | myDGR prediction |
| 345 | SRS015893_WUGC_scaffold_42257 | Human Metagenome | myDGR prediction |
| 346 | SRS015854_WUGC_scaffold_39752 | Human Metagenome | myDGR prediction |
| 347 | SRS015854_WUGC_scaffold_29429 | Human Metagenome | myDGR prediction |
| 348 | SRS015794_Baylor_scaffold_4524 | Human Metagenome | myDGR prediction |
| 349 | SRS015782_WUGC_scaffold_6228 | Human Metagenome | myDGR prediction |
| 350 | SRS015782_WUGC_scaffold_40748 | Human Metagenome | myDGR prediction |
| 351 | SRS015782_WUGC_scaffold_28901 | Human Metagenome | myDGR prediction |
| 352 | SRS015782_WUGC_scaffold_1599 | Human Metagenome | myDGR prediction |
| 353 | SRS015782_WUGC_scaffold_1357 | Human Metagenome | myDGR prediction |
| 354 | SRS015663_WUGC_scaffold_47728 | Human Metagenome | myDGR prediction |
| 355 | SRS015663_WUGC_scaffold_36939 | Human Metagenome | myDGR prediction |
| 356 | SRS015663_WUGC_scaffold_33235 | Human Metagenome | myDGR prediction |
| 357 | SRS015663_WUGC_scaffold_27774 | Human Metagenome | myDGR prediction |
| 358 | SRS015663_WUGC_scaffold_21269 | Human Metagenome | myDGR prediction |
| 359 | SRS015663_WUGC_scaffold_16190 | Human Metagenome | myDGR prediction |
| 360 | SRS015663_WUGC_scaffold_11604 | Human Metagenome | myDGR prediction |
| 361 | SRS015578_WUGC_scaffold_51480 | Human Metagenome | myDGR prediction |
| 362 | SRS015578_WUGC_scaffold_51233 | Human Metagenome | myDGR prediction |
| 363 | SRS015578_WUGC_scaffold_31117 | Human Metagenome | myDGR prediction |
| 364 | SRS015578_WUGC_scaffold_29761 | Human Metagenome | myDGR prediction |
| 365 | SRS015369_Baylor_scaffold_5833 | Human Metagenome | myDGR prediction |
| 366 | SRS015264_WUGC_scaffold_57793 | Human Metagenome | myDGR prediction |
| 367 | SRS015264_WUGC_scaffold_50181 | Human Metagenome | myDGR prediction |
| 368 | SRS015264_WUGC_scaffold_3386 | Human Metagenome | myDGR prediction |
| 369 | SRS015264_WUGC_scaffold_3108 | Human Metagenome | myDGR prediction |
| 370 | SRS015264_WUGC_scaffold_13311 | Human Metagenome | myDGR prediction |
| 371 | SRS015217_WUGC_scaffold_6606 | Human Metagenome | myDGR prediction |
| 372 | SRS015217_WUGC_scaffold_23239 | Human Metagenome | myDGR prediction |
| 373 | SRS015217_WUGC_scaffold_19551 | Human Metagenome | myDGR prediction |
| 374 | SRS015209_WUGC_scaffold_39092 | Human Metagenome | myDGR prediction |
| 375 | SRS015190_WUGC_scaffold_9866 | Human Metagenome | myDGR prediction |
| 376 | SRS015190_WUGC_scaffold_24113 | Human Metagenome | myDGR prediction |
| 377 | SRS015190_WUGC_scaffold_20240 | Human Metagenome | myDGR prediction |
| 378 | SRS015190_WUGC_scaffold_19745 | Human Metagenome | myDGR prediction |
| 379 | SRS015190_WUGC_scaffold_12656 | Human Metagenome | myDGR prediction |
| 380 | SRS015190_WUGC_scaffold_11551 | Human Metagenome | myDGR prediction |
| 381 | SRS015190_WUGC_scaffold_10956 | Human Metagenome | myDGR prediction |
| 382 | SRS015133_WUGC_scaffold_4781 | Human Metagenome | myDGR prediction |
| 383 | SRS015133_WUGC_scaffold_415 | Human Metagenome | myDGR prediction |
| 384 | SRS015133_WUGC_scaffold_38389 | Human Metagenome | myDGR prediction |
| 385 | SRS015133_WUGC_scaffold_33140 | Human Metagenome | myDGR prediction |
| 386 | SRS015133_WUGC_scaffold_12331 | Human Metagenome | myDGR prediction |
| 387 | SRS015133_WUGC_scaffold_11890 | Human Metagenome | myDGR prediction |
| 388 | SRS015065_WUGC_scaffold_9473 | Human Metagenome | myDGR prediction |
| 389 | SRS015065_WUGC_scaffold_2517 | Human Metagenome | myDGR prediction |
| 390 | SRS015065_WUGC_scaffold_12353 | Human Metagenome | myDGR prediction |
| 391 | SRS014979_WUGC_scaffold_5470 | Human Metagenome | myDGR prediction |
| 392 | SRS014979_WUGC_scaffold_34144 | Human Metagenome | myDGR prediction |
| 393 | SRS014979_WUGC_scaffold_31532 | Human Metagenome | myDGR prediction |
| 394 | SRS014979_WUGC_scaffold_10969 | Human Metagenome | myDGR prediction |
| 395 | SRS014923_WUGC_scaffold_59580 | Human Metagenome | myDGR prediction |
| 396 | SRS014923_WUGC_scaffold_58177 | Human Metagenome | myDGR prediction |
| 397 | SRS014923_WUGC_scaffold_56344 | Human Metagenome | myDGR prediction |
| 398 | SRS014923_WUGC_scaffold_5508 | Human Metagenome | myDGR prediction |
| 399 | SRS014923_WUGC_scaffold_51122 | Human Metagenome | myDGR prediction |
| 400 | SRS014923_WUGC_scaffold_28600 | Human Metagenome | myDGR prediction |
| 401 | SRS014923_WUGC_scaffold_17878 | Human Metagenome | myDGR prediction |
| 402 | SRS014923_WUGC_scaffold_16003 | Human Metagenome | myDGR prediction |
| 403 | SRS014923_WUGC_scaffold_15552 | Human Metagenome | myDGR prediction |
| 404 | SRS014923_WUGC_scaffold_13439 | Human Metagenome | myDGR prediction |
| 405 | SRS014683_WUGC_scaffold_50874 | Human Metagenome | myDGR prediction |
| 406 | SRS014683_WUGC_scaffold_22791 | Human Metagenome | myDGR prediction |
| 407 | SRS014613_WUGC_scaffold_9625 | Human Metagenome | myDGR prediction |
| 408 | SRS014613_WUGC_scaffold_3983 | Human Metagenome | myDGR prediction |
| 409 | SRS014613_WUGC_scaffold_3234 | Human Metagenome | myDGR prediction |
| 410 | SRS014613_WUGC_scaffold_15665 | Human Metagenome | myDGR prediction |
| 411 | SRS014613_WUGC_scaffold_13308 | Human Metagenome | myDGR prediction |
| 412 | SRS014459_WUGC_scaffold_93499 | Human Metagenome | myDGR prediction |
| 413 | SRS014459_WUGC_scaffold_93237 | Human Metagenome | myDGR prediction |
| 414 | SRS014459_WUGC_scaffold_86535 | Human Metagenome | myDGR prediction |
| 415 | SRS014459_WUGC_scaffold_77554 | Human Metagenome | myDGR prediction |
| 416 | SRS014459_WUGC_scaffold_65199 | Human Metagenome | myDGR prediction |
| 417 | SRS014459_WUGC_scaffold_47457 | Human Metagenome | myDGR prediction |
| 418 | SRS014459_WUGC_scaffold_39340 | Human Metagenome | myDGR prediction |
| 419 | SRS014459_WUGC_scaffold_35466 | Human Metagenome | myDGR prediction |
| 420 | SRS014459_WUGC_scaffold_31762 | Human Metagenome | myDGR prediction |
| 421 | SRS014459_WUGC_scaffold_27153 | Human Metagenome | myDGR prediction |
| 422 | SRS014459_WUGC_scaffold_24445 | Human Metagenome | myDGR prediction |
| 423 | SRS014459_WUGC_scaffold_24026 | Human Metagenome | myDGR prediction |
| 424 | SRS014459_WUGC_scaffold_21697 | Human Metagenome | myDGR prediction |
| 425 | SRS014459_WUGC_scaffold_17576 | Human Metagenome | myDGR prediction |
| 426 | SRS014313_WUGC_scaffold_3645 | Human Metagenome | myDGR prediction |
| 427 | SRS014287_WUGC_scaffold_39366 | Human Metagenome | myDGR prediction |
| 428 | SRS014287_WUGC_scaffold_38207 | Human Metagenome | myDGR prediction |
| 429 | SRS014287_WUGC_scaffold_21386 | Human Metagenome | myDGR prediction |
| 430 | SRS014287_WUGC_scaffold_11775 | Human Metagenome | myDGR prediction |
| 431 | SRS014287_WUGC_scaffold_10223 | Human Metagenome | myDGR prediction |
| 432 | SRS013951_WUGC_scaffold_8022 | Human Metagenome | myDGR prediction |
| 433 | SRS013951_WUGC_scaffold_69149 | Human Metagenome | myDGR prediction |
| 434 | SRS013951_WUGC_scaffold_69110 | Human Metagenome | myDGR prediction |
| 435 | SRS013951_WUGC_scaffold_64263 | Human Metagenome | myDGR prediction |
| 436 | SRS013951_WUGC_scaffold_62110 | Human Metagenome | myDGR prediction |
| 437 | SRS013951_WUGC_scaffold_59639 | Human Metagenome | myDGR prediction |
| 438 | SRS013951_WUGC_scaffold_59596 | Human Metagenome | myDGR prediction |
| 439 | SRS013951_WUGC_scaffold_5621 | Human Metagenome | myDGR prediction |
| 440 | SRS013951_WUGC_scaffold_47656 | Human Metagenome | myDGR prediction |
| 441 | SRS013951_WUGC_scaffold_47639 | Human Metagenome | myDGR prediction |
| 442 | SRS013951_WUGC_scaffold_37798 | Human Metagenome | myDGR prediction |
| 443 | SRS013951_WUGC_scaffold_15927 | Human Metagenome | myDGR prediction |
| 444 | SRS013951_WUGC_scaffold_14463 | Human Metagenome | myDGR prediction |
| 445 | SRS013800_Baylor_scaffold_9909 | Human Metagenome | myDGR prediction |
| 446 | SRS013800_Baylor_scaffold_15823 | Human Metagenome | myDGR prediction |
| 447 | SRS013687_Baylor_scaffold_9205 | Human Metagenome | myDGR prediction |
| 448 | SRS013687_Baylor_scaffold_59360 | Human Metagenome | myDGR prediction |
| 449 | SRS013687_Baylor_scaffold_56687 | Human Metagenome | myDGR prediction |
| 450 | SRS013687_Baylor_scaffold_54985 | Human Metagenome | myDGR prediction |
| 451 | SRS013687_Baylor_scaffold_43559 | Human Metagenome | myDGR prediction |
| 452 | SRS013687_Baylor_scaffold_43495 | Human Metagenome | myDGR prediction |
| 453 | SRS013687_Baylor_scaffold_31928 | Human Metagenome | myDGR prediction |
| 454 | SRS013687_Baylor_scaffold_21809 | Human Metagenome | myDGR prediction |
| 455 | SRS013687_Baylor_scaffold_21615 | Human Metagenome | myDGR prediction |
| 456 | SRS013687_Baylor_scaffold_14625 | Human Metagenome | myDGR prediction |
| 457 | SRS013542_LANL_scaffold_270 | Human Metagenome | myDGR prediction |
| 458 | SRS013521_LANL_scaffold_20661 | Human Metagenome | myDGR prediction |
| 459 | SRS013476_Baylor_scaffold_6811 | Human Metagenome | myDGR prediction |
| 460 | SRS013215_Baylor_scaffold_1384 | Human Metagenome | myDGR prediction |
| 461 | SRS012902_Baylor_scaffold_2704 | Human Metagenome | myDGR prediction |
| 462 | SRS012273_Baylor_scaffold_8590 | Human Metagenome | myDGR prediction |
| 463 | SRS012273_Baylor_scaffold_8246 | Human Metagenome | myDGR prediction |
| 464 | SRS012273_Baylor_scaffold_34301 | Human Metagenome | myDGR prediction |
| 465 | SRS011586_WUGC_scaffold_7434 | Human Metagenome | myDGR prediction |
| 466 | SRS011586_WUGC_scaffold_34656 | Human Metagenome | myDGR prediction |
| 467 | SRS011586_WUGC_scaffold_2044 | Human Metagenome | myDGR prediction |
| 468 | SRS011586_WUGC_scaffold_11330 | Human Metagenome | myDGR prediction |
| 469 | SRS011586_WUGC_scaffold_10335 | Human Metagenome | myDGR prediction |
| 470 | SRS011529_WUGC_scaffold_3649 | Human Metagenome | myDGR prediction |
| 471 | SRS011529_WUGC_scaffold_31780 | Human Metagenome | myDGR prediction |
| 472 | SRS011452_WUGC_scaffold_2829 | Human Metagenome | myDGR prediction |
| 473 | SRS011405_WUGC_scaffold_21229 | Human Metagenome | myDGR prediction |
| 474 | SRS011302_Baylor_scaffold_24381 | Human Metagenome | myDGR prediction |
| 475 | SRS011302_Baylor_scaffold_1538 | Human Metagenome | myDGR prediction |
| 476 | SRS011271_WUGC_scaffold_36635 | Human Metagenome | myDGR prediction |
| 477 | SRS011271_WUGC_scaffold_27850 | Human Metagenome | myDGR prediction |
| 478 | SRS011271_WUGC_scaffold_24007 | Human Metagenome | myDGR prediction |
| 479 | SRS011243_Baylor_scaffold_48301 | Human Metagenome | myDGR prediction |
| 480 | SRS011239_Baylor_scaffold_8091 | Human Metagenome | myDGR prediction |
| 481 | SRS011239_Baylor_scaffold_4178 | Human Metagenome | myDGR prediction |
| 482 | SRS011239_Baylor_scaffold_29381 | Human Metagenome | myDGR prediction |
| 483 | SRS011239_Baylor_scaffold_29301 | Human Metagenome | myDGR prediction |
| 484 | SRS011239_Baylor_scaffold_2051 | Human Metagenome | myDGR prediction |
| 485 | SRS011239_Baylor_scaffold_1978 | Human Metagenome | myDGR prediction |
| 486 | SRS011134_Baylor_scaffold_29667 | Human Metagenome | myDGR prediction |
| 487 | C5527333 | Human Metagenome | myDGR prediction |
| 488 | C5167117 | Human Metagenome | myDGR prediction |
| 489 | C5159013 | Human Metagenome | myDGR prediction |
| 490 | C4772571 | Human Metagenome | myDGR prediction |
| 491 | C4412323 | Human Metagenome | myDGR prediction |
| 492 | C4276967 | Human Metagenome | myDGR prediction |
| 493 | C4275891 | Human Metagenome | myDGR prediction |
| 494 | C4259500 | Human Metagenome | myDGR prediction |
| 495 | C4249812 | Human Metagenome | myDGR prediction |
| 496 | C3869178 | Human Metagenome | myDGR prediction |
| 497 | C3750933 | Human Metagenome | myDGR prediction |
| 498 | C3661977 | Human Metagenome | myDGR prediction |
| 499 | C3640976 | Human Metagenome | myDGR prediction |
| 500 | C3533957 | Human Metagenome | myDGR prediction |
| 501 | C3531371 | Human Metagenome | myDGR prediction |
| 502 | C3499983 | Human Metagenome | myDGR prediction |
| 503 | C3372742 | Human Metagenome | myDGR prediction |
| 504 | C3323548 | Human Metagenome | myDGR prediction |
| 505 | C3308112 | Human Metagenome | myDGR prediction |
| 506 | C3236958 | Human Metagenome | myDGR prediction |
| 507 | C3236803 | Human Metagenome | myDGR prediction |
| 508 | C3233957 | Human Metagenome | myDGR prediction |
| 509 | C3232672 | Human Metagenome | myDGR prediction |
| 510 | C3208701 | Human Metagenome | myDGR prediction |
| 511 | C3178453 | Human Metagenome | myDGR prediction |
| 512 | C3073331 | Human Metagenome | myDGR prediction |
| 513 | C3059246 | Human Metagenome | myDGR prediction |
| 514 | C3057496 | Human Metagenome | myDGR prediction |
| 515 | C2870985 | Human Metagenome | myDGR prediction |
| 516 | C2822214 | Human Metagenome | myDGR prediction |
| 517 | C2813255 | Human Metagenome | myDGR prediction |
| 518 | C2736404 | Human Metagenome | myDGR prediction |
| 519 | C2697151 | Human Metagenome | myDGR prediction |
| 520 | C2647608 | Human Metagenome | myDGR prediction |
| 521 | C2634073 | Human Metagenome | myDGR prediction |
| 522 | C2626114 | Human Metagenome | myDGR prediction |
| 523 | C2592563 | Human Metagenome | myDGR prediction |
| 524 | C2586223 | Human Metagenome | myDGR prediction |
| 525 | C2492782 | Human Metagenome | myDGR prediction |
| 526 | C2479546 | Human Metagenome | myDGR prediction |
| 527 | C2455298 | Human Metagenome | myDGR prediction |
| 528 | C2397089 | Human Metagenome | myDGR prediction |
| 529 | C2391720 | Human Metagenome | myDGR prediction |
| 530 | C2384524 | Human Metagenome | myDGR prediction |
| 531 | C2383034 | Human Metagenome | myDGR prediction |
| 532 | C2382976 | Human Metagenome | myDGR prediction |
| 533 | C2381144 | Human Metagenome | myDGR prediction |
| 534 | C2350512 | Human Metagenome | myDGR prediction |
| 535 | C2346318 | Human Metagenome | myDGR prediction |
| 536 | C2209444 | Human Metagenome | myDGR prediction |
| 537 | C2209264 | Human Metagenome | myDGR prediction |
| 538 | C2209030 | Human Metagenome | myDGR prediction |
| 539 | C2204788 | Human Metagenome | myDGR prediction |
| 540 | C2204032 | Human Metagenome | myDGR prediction |
| 541 | C2197865 | Human Metagenome | myDGR prediction |
| 542 | C2072421 | Human Metagenome | myDGR prediction |
| 543 | C2072169 | Human Metagenome | myDGR prediction |
| 544 | C1935518 | Human Metagenome | myDGR prediction |
| 545 | C1935188 | Human Metagenome | myDGR prediction |
| 546 | C1905524 | Human Metagenome | myDGR prediction |
| 547 | C1876891 | Human Metagenome | myDGR prediction |
| 548 | C1873301 | Human Metagenome | myDGR prediction |
| 549 | C1842429 | Human Metagenome | myDGR prediction |
| 550 | C1830124 | Human Metagenome | myDGR prediction |
| 551 | C1829670 | Human Metagenome | myDGR prediction |
| 552 | C1814024 | Human Metagenome | myDGR prediction |
| 553 | C1764656 | Human Metagenome | myDGR prediction |
| 554 | C1746428 | Human Metagenome | myDGR prediction |
| 555 | C1744576 | Human Metagenome | myDGR prediction |
| 556 | C1622866 | Human Metagenome | myDGR prediction |
| 557 | C1591216 | Human Metagenome | myDGR prediction |
| 558 | C1505893 | Human Metagenome | myDGR prediction |
| 559 | C1462390 | Human Metagenome | myDGR prediction |
Putative DGRs in human and animals gut metagenomes (ref)
| index | gut metagenome | myDGR results |
|---|---|---|
| 1 | ERR248260 Chicken | myDGR prediction |
| 2 | ERR248261 Cow | myDGR prediction |
| 3 | ERR248262 Human | myDGR prediction |
| 4 | ERR248263 Pig | myDGR prediction |
Putative DGRs in pig gut metagenomes (ref)
| index | Metagenome | myDGR results |
|---|---|---|
| 1 | ERR1135178 | myDGR prediction |
| 2 | ERR1135179 | myDGR prediction |
| 3 | ERR1135180 | myDGR prediction |
| 4 | ERR1135181 | myDGR prediction |